
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (11): 2510-2522.doi: 10.16420/j.issn.0513-353x.2024-0066
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
CHEN Fangce1, HU Pingzheng2, XIE Weiwei1, WANG Zhengpeng1, ZHONG Chuangnan1, LI Haiyan3, HE Yehua1, PENG Ze1, WAN Baoxiong3,*( ), LIU Chaoyang1,*(
), LIU Chaoyang1,*( )
)
Received:2024-04-02
Revised:2024-09-03
Online:2024-12-12
Published:2024-11-25
Contact:
WAN Baoxiong, LIU Chaoyang
CHEN Fangce, HU Pingzheng, XIE Weiwei, WANG Zhengpeng, ZHONG Chuangnan, LI Haiyan, HE Yehua, PENG Ze, WAN Baoxiong, LIU Chaoyang. Development and Application of Prunus salicina InDel Markers Based on Genome Re-sequencing[J]. Acta Horticulturae Sinica, 2024, 51(11): 2510-2522.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0066
| 类群 Group | 编号 Code | 样品名称 Sample name | 来源 Login | 果肉颜色 Flesh color |
|---|---|---|---|---|
| 华南品种 | GD01 | 灌村三华李Guancun Sanhuali | 广东Guangdong | 红色Red |
| Southern cultivar | GD02 | 上罗三华李Shangluo Sanhuali | 广东Guangdong | 红色Red |
| GD03 | 大蜜李Damili | 广东Guangdong | 红色Red | |
| GD04 | 华蜜大蜜李Huami Damili | 广东Guangdong | 红色Red | |
| GD05 | 白脆鸡麻李实生Baicuijimali Shisheng | 广东Guangdong | 黄色Yellow | |
| GD06 | 白脆鸡麻李Baicuijimali | 广东Guangdong | 红色Red | |
| GD07 | 竹丝李Zhusili | 广东Guangdong | 黄色Yellow | |
| GD08 | 硬枝李Yingzhili | 广东Guangdong | 黄色Yellow | |
| GD09 | 连麻三月李Lianma Sanyueli | 广东Guangdong | 黄色Yellow | |
| GD10 | 桂峰三月李Guifeng Sanyueli | 广东Guangdong | 黄色Yellow | |
| GD11 | 三月李实生Sanyueli Shisheng | 广东Guangdong | 黄色Yellow | |
| GD12 | 岭溪李实生Lingxili Shisheng | 广东Guangdong | 黄色Yellow | |
| GD13 | 乳源岭溪李Ruyuan Lingxili | 广东Guangdong | 黄色Yellow | |
| GD14 | 乳源大黄李Ruyuan Dahuangli | 广东Guangdong | 黄色Yellow | |
| GD15 | 乳源1号Ruyuan 1 | 广东Guangdong | 黄色Yellow | |
| GD16 | 早竹李Zaozhuli | 广东Guangdong | 黄色Yellow | |
| GD17 | 香蜜李Xiangmili | 广东Guangdong | 红色Red | |
| GD18 | 学佬李Xuelaoli | 广东Guangdong | 黄色Yellow | |
| GD19 | 瑶山李Yaoshanli | 广东Guangdong | 红色Red | |
| GD20 | 韶关4号野李Shaoguan 4 Yeli | 广东Guangdong | 黄色Yellow | |
| GD21 | 韶关6号野李Shaoguan 6 Yeli | 广东Guangdong | 黄色Yellow | |
| GD22 | 猪血李Zhuxueli | 广东Guangdong | 红色Red | |
| GD23 | 从早1号Congzao 1 | 广东Guangdong | 黄色Yellow | |
| GD24 | 云开1号Yunkai 1 | 广东Guangdong | 红色Red | |
| GD25 | 早禾李Zaoheli | 广东Guangdong | 黄色Yellow | |
| GD26 | 串珠李Chuanzhuli | 广东Guangdong | 黄色Yellow | |
| GD27 | 黄金㮈李Huangjin Naili | 广东Guangdong | 黄色Yellow | |
| GX01 | 乐业早熟李Leye Zaoshuli | 广西Guangxi | 黄色Yellow | |
| GX02 | 广西大水李Guangxi Dashuili | 广西Guangxi | 黄色Yellow | |
| GX03 | 胭脂李Yanzhili | 广西Guangxi | 红色Red | |
| GX04 | 苞谷李Baoguli | 广西Guangxi | 黄色Yellow | |
| GX05 | 凌云鸡血李Lingyun Jixueli | 广西Guangxi | 红色Red | |
| GX06 | 凌云桐壳李Lingyun Tongkeli | 广西Guangxi | 黄色Yellow | |
| GX07 | 乐业黄壳李Leye Huangkeli | 广西Guangxi | 黄色Yellow | |
| GX08 | 乐业小栽秧李Leye Xiaozaiyangli | 广西Guangxi | 黄色Yellow | |
| GX09 | 乐业红牛心李Leye Hongniuxinli | 广西Guangxi | 黄色Yellow | |
| GX10 | 化峒镇血红李Huadongzhen Xuehongli | 广西Guangxi | 红色Red | |
| GX11 | 乐业黄皮李Leye Huangpili | 广西Guangxi | 黄色Yellow | |
| GX12 | 乐业牛鸡心李Leye Niujixinli | 广西Guangxi | 黄色Yellow | |
| 西南品种 | GZ01 | 蜂糖李Fengtangli | 贵州Guizhou | 黄色Yellow |
| Southwest cultivar | GZ02 | 一点红蜂糖李Yidianhong Fengtangli | 贵州Guizhou | 黄色Yellow |
| GZ03 | 蜜李1号Mili 1 | 贵州Guizhou | 黄色Yellow | |
| GZ04 | 安顺本地晚熟李Anshun Bendi Wanshuli | 贵州Guizhou | 黄色Yellow | |
| GZ05 | 四月李Siyueli | 贵州Guizhou | 黄色Yellow | |
| GZ06 | 打帮李Dabangli | 贵州Guizhou | 黄色Yellow | |
| GZ07 | 冰脆李Bingcuili | 贵州Guizhou | 黄色Yellow | |
| GZ08 | 棉花李Mianhuali | 贵州Guizhou | 黄色Yellow | |
| GZ09 | 紫云本地红李Ziyun Bendi Hongli | 贵州Guizhou | 黄色Yellow | |
| GZ10 | 脆红李Cuihongli | 贵州Guizhou | 黄色Yellow | |
| GZ11 | 沿河空心李Yanhe Kongxinli | 贵州Guizhou | 黄色Yellow | |
| GZ12 | 黄蜡李Huanglali | 贵州Guizhou | 黄色Yellow | |
| GZ13 | 九阡李Jiuqianli | 贵州Guizhou | 黄绿色Chartreuse | |
| SC01 | 晚熟青脆李Wanshu Qingcuili | 四川Sichuan | 黄色Yellow | |
| SC02 | 五月脆Wuyuecui | 四川Sichuan | 黄绿色Chartreuse | |
| SC03 | 苍溪早青脆Cangxi Zaoqingcui | 四川Sichuan | 黄色Yellow | |
| SC04 | 江安李Jianganli | 四川Sichuan | 黄绿色Chartreuse | |
| CQ01 | 铜梁龙李Tongliang Longli | 重庆Chongqing | 黄色Yellow | |
| CQ02 | 粉黛脆李Fendai Cuili | 重庆Chongqing | 黄色Yellow | |
| YN01 | 云南黄李Yunnan Huangli | 云南Yunnan | 黄色Yellow | |
| 南方其他地区品种 | HB01 | 五峰翰林空心李Wufeng Hanlin Kongxinli | 湖北Hubei | 黄色Yellow |
| Other southern cultivar | HB02 | 油头青Youtouqing | 湖北Hubei | 黄色Yellow |
| ZJ01 | 桃形李Taoxingli | 浙江Zhejiang | 黄色Yellow | |
| ZJ02 | 檇李Zuili | 浙江Zhejiang | 黄色yellow | |
| ZJ03 | 金塘李Jintangli | 浙江Zhejiang | 红色Red | |
| FJ01 | 芙蓉李Furongli | 福建Fujian | 红色Red | |
| FJ02 | 油奈Younai | 福建Fujian | 黄色Yellow | |
| FJ03 | 鹅黄李Ehuangli | 福建Fujian | 黄色Yellow | |
| 北方品种 | SD01 | 山东早红Shandong Zaohong | 山东Shandong | 黄色Yellow |
| Northern cultivar | SD02 | 玉皇李Yuhuangli | 山东Shandong | 黄色Yellow |
| HN01 | 灰子李Huizili | 河南Henan | 黄色Yellow | |
| XJ01 | 新疆黄李Xinjiang Huangli | 新疆Xinjiang | 黄色Yellow | |
| SX01 | 商县梅李Shangxian Meili | 陕西Shaanxi | 黄色Yellow | |
| LN01 | 大红袍Dahongpao | 辽宁Liaoning | 黄色Yellow | |
| LN02 | 国峰7号Guofeng 7 | 辽宁Liaoning | 黄色Yellow | |
| JL01 | 晚金玉Wanjinyu | 吉林Jinlin | 黄色Yellow | |
| JL02 | 松祥桃李Songxiang Taoli | 吉林Jinlin | 黄色Yellow | |
| HLJ01 | 龙园蜜李Longyuan Mili | 黑龙江Heilongjiang | 黄色Yellow | |
| HLJ02 | 秋甜李Qiutianli | 黑龙江Heilongjiang | 黄色Yellow | |
| 杏李品种P. simonii | XL01 | 味帝Weidi | 国外Foreign | 红色Red |
Table 1 Information of 79 Prunus cultivars
| 类群 Group | 编号 Code | 样品名称 Sample name | 来源 Login | 果肉颜色 Flesh color |
|---|---|---|---|---|
| 华南品种 | GD01 | 灌村三华李Guancun Sanhuali | 广东Guangdong | 红色Red |
| Southern cultivar | GD02 | 上罗三华李Shangluo Sanhuali | 广东Guangdong | 红色Red |
| GD03 | 大蜜李Damili | 广东Guangdong | 红色Red | |
| GD04 | 华蜜大蜜李Huami Damili | 广东Guangdong | 红色Red | |
| GD05 | 白脆鸡麻李实生Baicuijimali Shisheng | 广东Guangdong | 黄色Yellow | |
| GD06 | 白脆鸡麻李Baicuijimali | 广东Guangdong | 红色Red | |
| GD07 | 竹丝李Zhusili | 广东Guangdong | 黄色Yellow | |
| GD08 | 硬枝李Yingzhili | 广东Guangdong | 黄色Yellow | |
| GD09 | 连麻三月李Lianma Sanyueli | 广东Guangdong | 黄色Yellow | |
| GD10 | 桂峰三月李Guifeng Sanyueli | 广东Guangdong | 黄色Yellow | |
| GD11 | 三月李实生Sanyueli Shisheng | 广东Guangdong | 黄色Yellow | |
| GD12 | 岭溪李实生Lingxili Shisheng | 广东Guangdong | 黄色Yellow | |
| GD13 | 乳源岭溪李Ruyuan Lingxili | 广东Guangdong | 黄色Yellow | |
| GD14 | 乳源大黄李Ruyuan Dahuangli | 广东Guangdong | 黄色Yellow | |
| GD15 | 乳源1号Ruyuan 1 | 广东Guangdong | 黄色Yellow | |
| GD16 | 早竹李Zaozhuli | 广东Guangdong | 黄色Yellow | |
| GD17 | 香蜜李Xiangmili | 广东Guangdong | 红色Red | |
| GD18 | 学佬李Xuelaoli | 广东Guangdong | 黄色Yellow | |
| GD19 | 瑶山李Yaoshanli | 广东Guangdong | 红色Red | |
| GD20 | 韶关4号野李Shaoguan 4 Yeli | 广东Guangdong | 黄色Yellow | |
| GD21 | 韶关6号野李Shaoguan 6 Yeli | 广东Guangdong | 黄色Yellow | |
| GD22 | 猪血李Zhuxueli | 广东Guangdong | 红色Red | |
| GD23 | 从早1号Congzao 1 | 广东Guangdong | 黄色Yellow | |
| GD24 | 云开1号Yunkai 1 | 广东Guangdong | 红色Red | |
| GD25 | 早禾李Zaoheli | 广东Guangdong | 黄色Yellow | |
| GD26 | 串珠李Chuanzhuli | 广东Guangdong | 黄色Yellow | |
| GD27 | 黄金㮈李Huangjin Naili | 广东Guangdong | 黄色Yellow | |
| GX01 | 乐业早熟李Leye Zaoshuli | 广西Guangxi | 黄色Yellow | |
| GX02 | 广西大水李Guangxi Dashuili | 广西Guangxi | 黄色Yellow | |
| GX03 | 胭脂李Yanzhili | 广西Guangxi | 红色Red | |
| GX04 | 苞谷李Baoguli | 广西Guangxi | 黄色Yellow | |
| GX05 | 凌云鸡血李Lingyun Jixueli | 广西Guangxi | 红色Red | |
| GX06 | 凌云桐壳李Lingyun Tongkeli | 广西Guangxi | 黄色Yellow | |
| GX07 | 乐业黄壳李Leye Huangkeli | 广西Guangxi | 黄色Yellow | |
| GX08 | 乐业小栽秧李Leye Xiaozaiyangli | 广西Guangxi | 黄色Yellow | |
| GX09 | 乐业红牛心李Leye Hongniuxinli | 广西Guangxi | 黄色Yellow | |
| GX10 | 化峒镇血红李Huadongzhen Xuehongli | 广西Guangxi | 红色Red | |
| GX11 | 乐业黄皮李Leye Huangpili | 广西Guangxi | 黄色Yellow | |
| GX12 | 乐业牛鸡心李Leye Niujixinli | 广西Guangxi | 黄色Yellow | |
| 西南品种 | GZ01 | 蜂糖李Fengtangli | 贵州Guizhou | 黄色Yellow |
| Southwest cultivar | GZ02 | 一点红蜂糖李Yidianhong Fengtangli | 贵州Guizhou | 黄色Yellow |
| GZ03 | 蜜李1号Mili 1 | 贵州Guizhou | 黄色Yellow | |
| GZ04 | 安顺本地晚熟李Anshun Bendi Wanshuli | 贵州Guizhou | 黄色Yellow | |
| GZ05 | 四月李Siyueli | 贵州Guizhou | 黄色Yellow | |
| GZ06 | 打帮李Dabangli | 贵州Guizhou | 黄色Yellow | |
| GZ07 | 冰脆李Bingcuili | 贵州Guizhou | 黄色Yellow | |
| GZ08 | 棉花李Mianhuali | 贵州Guizhou | 黄色Yellow | |
| GZ09 | 紫云本地红李Ziyun Bendi Hongli | 贵州Guizhou | 黄色Yellow | |
| GZ10 | 脆红李Cuihongli | 贵州Guizhou | 黄色Yellow | |
| GZ11 | 沿河空心李Yanhe Kongxinli | 贵州Guizhou | 黄色Yellow | |
| GZ12 | 黄蜡李Huanglali | 贵州Guizhou | 黄色Yellow | |
| GZ13 | 九阡李Jiuqianli | 贵州Guizhou | 黄绿色Chartreuse | |
| SC01 | 晚熟青脆李Wanshu Qingcuili | 四川Sichuan | 黄色Yellow | |
| SC02 | 五月脆Wuyuecui | 四川Sichuan | 黄绿色Chartreuse | |
| SC03 | 苍溪早青脆Cangxi Zaoqingcui | 四川Sichuan | 黄色Yellow | |
| SC04 | 江安李Jianganli | 四川Sichuan | 黄绿色Chartreuse | |
| CQ01 | 铜梁龙李Tongliang Longli | 重庆Chongqing | 黄色Yellow | |
| CQ02 | 粉黛脆李Fendai Cuili | 重庆Chongqing | 黄色Yellow | |
| YN01 | 云南黄李Yunnan Huangli | 云南Yunnan | 黄色Yellow | |
| 南方其他地区品种 | HB01 | 五峰翰林空心李Wufeng Hanlin Kongxinli | 湖北Hubei | 黄色Yellow |
| Other southern cultivar | HB02 | 油头青Youtouqing | 湖北Hubei | 黄色Yellow |
| ZJ01 | 桃形李Taoxingli | 浙江Zhejiang | 黄色Yellow | |
| ZJ02 | 檇李Zuili | 浙江Zhejiang | 黄色yellow | |
| ZJ03 | 金塘李Jintangli | 浙江Zhejiang | 红色Red | |
| FJ01 | 芙蓉李Furongli | 福建Fujian | 红色Red | |
| FJ02 | 油奈Younai | 福建Fujian | 黄色Yellow | |
| FJ03 | 鹅黄李Ehuangli | 福建Fujian | 黄色Yellow | |
| 北方品种 | SD01 | 山东早红Shandong Zaohong | 山东Shandong | 黄色Yellow |
| Northern cultivar | SD02 | 玉皇李Yuhuangli | 山东Shandong | 黄色Yellow |
| HN01 | 灰子李Huizili | 河南Henan | 黄色Yellow | |
| XJ01 | 新疆黄李Xinjiang Huangli | 新疆Xinjiang | 黄色Yellow | |
| SX01 | 商县梅李Shangxian Meili | 陕西Shaanxi | 黄色Yellow | |
| LN01 | 大红袍Dahongpao | 辽宁Liaoning | 黄色Yellow | |
| LN02 | 国峰7号Guofeng 7 | 辽宁Liaoning | 黄色Yellow | |
| JL01 | 晚金玉Wanjinyu | 吉林Jinlin | 黄色Yellow | |
| JL02 | 松祥桃李Songxiang Taoli | 吉林Jinlin | 黄色Yellow | |
| HLJ01 | 龙园蜜李Longyuan Mili | 黑龙江Heilongjiang | 黄色Yellow | |
| HLJ02 | 秋甜李Qiutianli | 黑龙江Heilongjiang | 黄色Yellow | |
| 杏李品种P. simonii | XL01 | 味帝Weidi | 国外Foreign | 红色Red |
| 染色体Chromosome | 插入Insertion | 缺失Deletion | 总计Total | InDel/% |
|---|---|---|---|---|
| Chr1 | 14 749 | 28 997 | 43 746 | 20.79 |
| Chr2 | 10 058 | 18 077 | 28 135 | 13.37 |
| Chr3 | 8 631 | 16 786 | 25 417 | 12.08 |
| Chr4 | 8 632 | 16 391 | 25 023 | 11.89 |
| Chr5 | 5 972 | 11 660 | 17 632 | 8.38 |
| Chr6 | 9 103 | 17 140 | 26 243 | 12.47 |
| Chr7 | 6 909 | 14 076 | 20 985 | 9.97 |
| Chr8 | 7 302 | 14 492 | 21 794 | 10.36 |
| 染色体支架Scaffolds | 526 | 931 | 1 457 | 0.69 |
| 总计Total | 71 882 | 138 550 | 210 432 | 100.00 |
Table 2 Whole-genome InDel locus statistics of plum
| 染色体Chromosome | 插入Insertion | 缺失Deletion | 总计Total | InDel/% |
|---|---|---|---|---|
| Chr1 | 14 749 | 28 997 | 43 746 | 20.79 |
| Chr2 | 10 058 | 18 077 | 28 135 | 13.37 |
| Chr3 | 8 631 | 16 786 | 25 417 | 12.08 |
| Chr4 | 8 632 | 16 391 | 25 023 | 11.89 |
| Chr5 | 5 972 | 11 660 | 17 632 | 8.38 |
| Chr6 | 9 103 | 17 140 | 26 243 | 12.47 |
| Chr7 | 6 909 | 14 076 | 20 985 | 9.97 |
| Chr8 | 7 302 | 14 492 | 21 794 | 10.36 |
| 染色体支架Scaffolds | 526 | 931 | 1 457 | 0.69 |
| 总计Total | 71 882 | 138 550 | 210 432 | 100.00 |
| 染色体 Chromosome | 引物名 Primer name | 物理位置 Physical location | 正向引物(5′-3′) Forward primer sequence | 反向引物(5′-3′) Reverse primer sequence |
|---|---|---|---|---|
| Chr1 | M1 | 38894318 | GGGGAGATGGAAAGTTAA | GTATTGCTCTTGGGCTGA |
| M2 | 47980202 | GGTTTTCTCGAAGTCGGTGAT | GGCTTGTGATATATTAGAACCCATG | |
| M3 | 48304762 | GGACCCCATGCTGAAGAAAA | CAAATCTAAGGGCAAGTCCAAT | |
| M4 | 48570078 | AGAAACCCACTATGTCACTG | AGACGGAGCATCAAATCA | |
| M5 | 49049772 | ATCCCGTCAAACTCACTA | TGGATTGAACCCATACAC | |
| M6 | 50387422 | CCACTTGGTCAAGGTTAAATGTC | GCAAATTGTAAGGGGTATGGTG | |
| M7 | 52179403 | TGAAGTAGTCAGGACCTCTGGC | GGTCAGAGGTGAGAGCGATGA | |
| M8 | 52719011 | TGGCTTCCAAACATCCTC | GTAGAGGCATCCAGCACA | |
| Chr2 | M9 | 12500445 | CAGGAGTAGGCTCAAGAT | TTATGATGGATGGGTATG |
| M10 | 31689717 | CGTCGTTAGCGAGTGAGT | TGGCACGAAACAAGCAGT | |
| M11 | 31757195 | GAGGCTCAATCGTAGACA | GATTCACGTTTGGGTTGT | |
| M12 | 33259045 | CCTGGAACCCACGAATTTCT | CTTTCCTGACCAAGACAGTGTGA | |
| M13 | 33866248 | CTCTAGGGTTCTTCTCTGCTATCTTC | GCTCGTTTGGTACGTTGGACT | |
| Chr3 | M14 | 1698465 | CACCCACTAAGACTGACG | CCTTTGCGGAACACTTTA |
| M15 | 6074639 | TAGAAACGCTACGAAGGC | AATGGCTAAATGCTTGGTT | |
| M16 | 6079326 | TTTGGGAAGTTAGTGGGA | TGCGGTCTTTCTCAACTCG | |
| M17 | 6982515 | TCTGGGAACCTGGTGTTA | AATGTGCCTTTCTGTCCC | |
| M18 | 27839434 | GGGTGGTCTGGTTCCCATAA | CATCGGCACAATAGTAGAGAAGC | |
| M19 | 28695379 | ACAACGAAGCCTCCTCTTGAG | CACCTTTCCCCACAAATTTTG | |
| M20 | 29098986 | ACCTTCCTCCTCATCATTGGA | ATAGTCGGGCTTTGCTGCTT | |
| M21 | 29210781 | TGATCCACACTTCCCTATGACAG | GCCACGTGCATTATTTCTAAGAG | |
| M22 | 29355454 | GCACGCACCCTTTCTTTTCT | CAGGGGCTTAGACAACCATCA | |
| M23 | 31644114 | CTTACTGCAGAAGTTCGTGAGAAA | CACGCCTTTTCTGTTTCTTCTT | |
| Chr4 | M24 | 7678285 | AGCCTGGTTGATTGTTGG | CAAACGCACTACGCAAAC |
| M25 | 7903736 | ACATCATCACGACCAGTA | CATCCACCAACAGCAACA | |
| M26 | 8483245 | CTGTCTTCCTCTGCCATTT | GAATCTGCTCGGTCAAAG | |
| M27 | 11530071 | ACCACCATTTTGCCAAGAGG | CCCAATCTTAGACTGGCCGTA | |
| M28 | 18058101 | CGCGCGGCTATACTATTTTT | GATTTGGGCAACCTCTACCC | |
| M29 | 19467756 | GGAGACCTTGCGACTGAT | AAGAGGTGGCTTGGTCAG | |
| M30 | 22025126 | CCAGCTCACATGCTCTTTTCC | GATGCCTCTCATATGCCACTTG | |
| M31 | 23776947 | AGATGTTGCCGAGAAAGG | CTCTATTTGCACCCACCC | |
| M32 | 24553291 | ACATAGGTGGCTGTGATTACGC | CATGTGATGGAGGAAGTCTTGC | |
| M33 | 31285226 | TGGCACACTACAATAGCACAATG | CCCAGGTCCCTCAACAGAAT | |
| Chr5 | M34 | 330880 | TTTAGCCAAGTGGTGTTT | GATGGCTACTGAGGAAGG |
| M35 | 7534231 | AAAGAACTTACTTGGGAGAT | CAGAATCCACCAAGAAGA | |
| M36 | 12530378 | GCAGTGGCGGAGCTAGAGT | TTTGCTAAGATACGATTAC | |
| M37 | 12533577 | GTATCAATACCCTTTGGC | GGCACATTTTGTCTCGGT | |
| M38 | 14348889 | CATTCCGTGAACCAAACAGG | CAATGCTGCCCAAATTGCTA | |
| M39 | 15641569 | CAACAGTCACCCCGCAATTA | GTCAGCATCCCATCCCAAAT | |
| M40 | 22769129 | CCAAAACTTTTCCCCAGAAAAC | GAGCTTGAGCAAGCATCGAG | |
| Chr6 | M41 | 3392066 | AAACTGACTCGGAAAGGT | TGCCATTGGTTCCCTACT |
| M42 | 3650929 | AATGAGCACGGGCAGATA | GATTGATAGCCTCGGTTG | |
| M43 | 8649659 | TATAGGGCACCTTCATCG | ACTGCTTTCAACTGTGGG | |
| M44 | 34659045 | TGCGACTCATCAGTTTGT | CCTTTCCCGTTCTACCAT | |
| Chr7 | M45 | 2356604 | CAAGGAGAAATAGAAGCGAAGA | CCCAAAGCACTCCGAAAG |
| M46 | 8337435 | CGGAAGAGGAGATGGTGGAA | GAGACTCTCACAGAGGCATTTCA | |
| M47 | 9880809 | ATGTTCCCTCCACTCCTT | AAGCCTTGTTGAGGTGAT | |
| M48 | 9973675 | CCTTGAAAAGACAGTGAACAGCA | TCCAAAGCCACCATTGTAACC | |
| M49 | 12448075 | CCATGAGGAGGGAAAACAAAA | AATGAGCGTGAGTATACAATTAGGC | |
| Chr8 | M50 | 5495485 | AAACTGACTCGGAAAGGT | ATTGCCATTGGTTCCCTA |
| M51 | 13608340 | TTCAGTGCCACAACAATAGGG | CGACGTACCCTTGTATCCCTTA | |
| M52 | 19291686 | ACATTGTTTGCTGGGCTAT | TCCCGTTTGACAACCATT | |
| M53 | 19739188 | AAACGCAAAGGACAAGAT | CGGGCAGTTCAAGGATTA |
Table 3 Primer sequence of 53 pairs of InDel markers
| 染色体 Chromosome | 引物名 Primer name | 物理位置 Physical location | 正向引物(5′-3′) Forward primer sequence | 反向引物(5′-3′) Reverse primer sequence |
|---|---|---|---|---|
| Chr1 | M1 | 38894318 | GGGGAGATGGAAAGTTAA | GTATTGCTCTTGGGCTGA |
| M2 | 47980202 | GGTTTTCTCGAAGTCGGTGAT | GGCTTGTGATATATTAGAACCCATG | |
| M3 | 48304762 | GGACCCCATGCTGAAGAAAA | CAAATCTAAGGGCAAGTCCAAT | |
| M4 | 48570078 | AGAAACCCACTATGTCACTG | AGACGGAGCATCAAATCA | |
| M5 | 49049772 | ATCCCGTCAAACTCACTA | TGGATTGAACCCATACAC | |
| M6 | 50387422 | CCACTTGGTCAAGGTTAAATGTC | GCAAATTGTAAGGGGTATGGTG | |
| M7 | 52179403 | TGAAGTAGTCAGGACCTCTGGC | GGTCAGAGGTGAGAGCGATGA | |
| M8 | 52719011 | TGGCTTCCAAACATCCTC | GTAGAGGCATCCAGCACA | |
| Chr2 | M9 | 12500445 | CAGGAGTAGGCTCAAGAT | TTATGATGGATGGGTATG |
| M10 | 31689717 | CGTCGTTAGCGAGTGAGT | TGGCACGAAACAAGCAGT | |
| M11 | 31757195 | GAGGCTCAATCGTAGACA | GATTCACGTTTGGGTTGT | |
| M12 | 33259045 | CCTGGAACCCACGAATTTCT | CTTTCCTGACCAAGACAGTGTGA | |
| M13 | 33866248 | CTCTAGGGTTCTTCTCTGCTATCTTC | GCTCGTTTGGTACGTTGGACT | |
| Chr3 | M14 | 1698465 | CACCCACTAAGACTGACG | CCTTTGCGGAACACTTTA |
| M15 | 6074639 | TAGAAACGCTACGAAGGC | AATGGCTAAATGCTTGGTT | |
| M16 | 6079326 | TTTGGGAAGTTAGTGGGA | TGCGGTCTTTCTCAACTCG | |
| M17 | 6982515 | TCTGGGAACCTGGTGTTA | AATGTGCCTTTCTGTCCC | |
| M18 | 27839434 | GGGTGGTCTGGTTCCCATAA | CATCGGCACAATAGTAGAGAAGC | |
| M19 | 28695379 | ACAACGAAGCCTCCTCTTGAG | CACCTTTCCCCACAAATTTTG | |
| M20 | 29098986 | ACCTTCCTCCTCATCATTGGA | ATAGTCGGGCTTTGCTGCTT | |
| M21 | 29210781 | TGATCCACACTTCCCTATGACAG | GCCACGTGCATTATTTCTAAGAG | |
| M22 | 29355454 | GCACGCACCCTTTCTTTTCT | CAGGGGCTTAGACAACCATCA | |
| M23 | 31644114 | CTTACTGCAGAAGTTCGTGAGAAA | CACGCCTTTTCTGTTTCTTCTT | |
| Chr4 | M24 | 7678285 | AGCCTGGTTGATTGTTGG | CAAACGCACTACGCAAAC |
| M25 | 7903736 | ACATCATCACGACCAGTA | CATCCACCAACAGCAACA | |
| M26 | 8483245 | CTGTCTTCCTCTGCCATTT | GAATCTGCTCGGTCAAAG | |
| M27 | 11530071 | ACCACCATTTTGCCAAGAGG | CCCAATCTTAGACTGGCCGTA | |
| M28 | 18058101 | CGCGCGGCTATACTATTTTT | GATTTGGGCAACCTCTACCC | |
| M29 | 19467756 | GGAGACCTTGCGACTGAT | AAGAGGTGGCTTGGTCAG | |
| M30 | 22025126 | CCAGCTCACATGCTCTTTTCC | GATGCCTCTCATATGCCACTTG | |
| M31 | 23776947 | AGATGTTGCCGAGAAAGG | CTCTATTTGCACCCACCC | |
| M32 | 24553291 | ACATAGGTGGCTGTGATTACGC | CATGTGATGGAGGAAGTCTTGC | |
| M33 | 31285226 | TGGCACACTACAATAGCACAATG | CCCAGGTCCCTCAACAGAAT | |
| Chr5 | M34 | 330880 | TTTAGCCAAGTGGTGTTT | GATGGCTACTGAGGAAGG |
| M35 | 7534231 | AAAGAACTTACTTGGGAGAT | CAGAATCCACCAAGAAGA | |
| M36 | 12530378 | GCAGTGGCGGAGCTAGAGT | TTTGCTAAGATACGATTAC | |
| M37 | 12533577 | GTATCAATACCCTTTGGC | GGCACATTTTGTCTCGGT | |
| M38 | 14348889 | CATTCCGTGAACCAAACAGG | CAATGCTGCCCAAATTGCTA | |
| M39 | 15641569 | CAACAGTCACCCCGCAATTA | GTCAGCATCCCATCCCAAAT | |
| M40 | 22769129 | CCAAAACTTTTCCCCAGAAAAC | GAGCTTGAGCAAGCATCGAG | |
| Chr6 | M41 | 3392066 | AAACTGACTCGGAAAGGT | TGCCATTGGTTCCCTACT |
| M42 | 3650929 | AATGAGCACGGGCAGATA | GATTGATAGCCTCGGTTG | |
| M43 | 8649659 | TATAGGGCACCTTCATCG | ACTGCTTTCAACTGTGGG | |
| M44 | 34659045 | TGCGACTCATCAGTTTGT | CCTTTCCCGTTCTACCAT | |
| Chr7 | M45 | 2356604 | CAAGGAGAAATAGAAGCGAAGA | CCCAAAGCACTCCGAAAG |
| M46 | 8337435 | CGGAAGAGGAGATGGTGGAA | GAGACTCTCACAGAGGCATTTCA | |
| M47 | 9880809 | ATGTTCCCTCCACTCCTT | AAGCCTTGTTGAGGTGAT | |
| M48 | 9973675 | CCTTGAAAAGACAGTGAACAGCA | TCCAAAGCCACCATTGTAACC | |
| M49 | 12448075 | CCATGAGGAGGGAAAACAAAA | AATGAGCGTGAGTATACAATTAGGC | |
| Chr8 | M50 | 5495485 | AAACTGACTCGGAAAGGT | ATTGCCATTGGTTCCCTA |
| M51 | 13608340 | TTCAGTGCCACAACAATAGGG | CGACGTACCCTTGTATCCCTTA | |
| M52 | 19291686 | ACATTGTTTGCTGGGCTAT | TCCCGTTTGACAACCATT | |
| M53 | 19739188 | AAACGCAAAGGACAAGAT | CGGGCAGTTCAAGGATTA |
| 标记名称Marker name | 主等位基因频率Major allele frquency | 基因型数Genotype number | Na 等位基因数 Number of allele | Hs 基因多样性指数 Gene diversity index | He 期望杂合度 Expected heterozygosity | PIC 多态性信息含量 Polymorphic information content |
|---|---|---|---|---|---|---|
| M1 | 0.8101 | 3 | 2 | 0.3076 | 0.3038 | 0.2603 |
| M2 | 0.8734 | 3 | 2 | 0.2211 | 0.1013 | 0.1967 |
| M3 | 0.7278 | 3 | 2 | 0.3962 | 0.1646 | 0.3177 |
| M4 | 0.8354 | 2 | 2 | 0.2750 | 0.3291 | 0.2372 |
| M5 | 0.6519 | 3 | 2 | 0.4539 | 0.443 | 0.3509 |
| M6 | 0.6582 | 3 | 2 | 0.4499 | 0.5063 | 0.3487 |
| M7 | 0.6282 | 3 | 2 | 0.4671 | 0.3333 | 0.3580 |
| M8 | 0.7025 | 3 | 2 | 0.4180 | 0.5190 | 0.3306 |
| M9 | 0.4808 | 6 | 3 | 0.6335 | 0.5513 | 0.5620 |
| M10 | 0.4286 | 3 | 3 | 0.6208 | 0 | 0.5404 |
| M11 | 0.5321 | 3 | 2 | 0.4979 | 0.5769 | 0.3740 |
| M12 | 0.6329 | 3 | 2 | 0.4647 | 0.5316 | 0.3567 |
| M13 | 0.5506 | 3 | 2 | 0.4949 | 0.7215 | 0.3724 |
| M14 | 0.5886 | 3 | 2 | 0.4843 | 0.6456 | 0.3670 |
| M15 | 0.6042 | 3 | 2 | 0.4783 | 0.2639 | 0.3639 |
| M16 | 0.4464 | 6 | 3 | 0.6435 | 0.2679 | 0.5697 |
| M17 | 0.3667 | 8 | 4 | 0.7212 | 0.4889 | 0.6703 |
| M18 | 0.5506 | 3 | 2 | 0.4949 | 0.2405 | 0.3724 |
| M19 | 0.5000 | 3 | 2 | 0.5000 | 0.3333 | 0.3750 |
| M20 | 0.8671 | 2 | 2 | 0.2305 | 0.2658 | 0.2039 |
| M21 | 0.8101 | 2 | 2 | 0.3076 | 0.3797 | 0.2603 |
| M22 | 0.5513 | 3 | 2 | 0.4947 | 0.2564 | 0.3724 |
| M23 | 0.9679 | 3 | 2 | 0.0620 | 0.0385 | 0.0601 |
| M24 | 0.6456 | 5 | 4 | 0.5042 | 0.5696 | 0.4375 |
| M25 | 0.5064 | 3 | 2 | 0.4999 | 0.8077 | 0.3750 |
| M26 | 0.5949 | 2 | 2 | 0.4820 | 0.8101 | 0.3658 |
| M27 | 0.5506 | 3 | 2 | 0.4949 | 0.5696 | 0.3724 |
| M28 | 0.5972 | 3 | 2 | 0.4811 | 0.4167 | 0.3654 |
| M29 | 0.7500 | 3 | 2 | 0.3750 | 0.2917 | 0.3047 |
| M30 | 0.7278 | 3 | 2 | 0.3962 | 0.2911 | 0.3177 |
| M31 | 0.5789 | 3 | 2 | 0.4875 | 0.1579 | 0.3687 |
| M32 | 0.6753 | 3 | 2 | 0.4385 | 0.2078 | 0.3424 |
| M33 | 0.7756 | 3 | 2 | 0.3480 | 0.3974 | 0.2875 |
| M34 | 0.6731 | 3 | 2 | 0.4401 | 0.2949 | 0.3432 |
| M35 | 0.4333 | 4 | 3 | 0.6516 | 0.0222 | 0.5784 |
| M36 | 0.8924 | 3 | 2 | 0.1920 | 0.1139 | 0.1736 |
| M37 | 0.9026 | 3 | 2 | 0.1758 | 0.1429 | 0.1604 |
| M38 | 0.6899 | 4 | 3 | 0.4423 | 0.3671 | 0.3644 |
| M39 | 0.8103 | 3 | 2 | 0.3074 | 0.1034 | 0.2601 |
| M40 | 0.8291 | 3 | 2 | 0.2834 | 0.3165 | 0.2432 |
| M41 | 0.6218 | 3 | 2 | 0.4703 | 0.5513 | 0.3597 |
| M42 | 0.6866 | 4 | 3 | 0.4740 | 0.0896 | 0.4216 |
| M43 | 0.6014 | 3 | 2 | 0.4794 | 0.1014 | 0.3645 |
| M44 | 0.7722 | 3 | 2 | 0.3519 | 0.3797 | 0.2900 |
| M45 | 0.8734 | 3 | 2 | 0.2211 | 0.2025 | 0.1967 |
| M46 | 0.7975 | 3 | 2 | 0.3230 | 0.3797 | 0.2709 |
| M47 | 0.6392 | 2 | 2 | 0.4612 | 0.7215 | 0.3549 |
| M48 | 0.5294 | 3 | 2 | 0.4983 | 0.1569 | 0.3741 |
| M49 | 0.6582 | 3 | 2 | 0.4499 | 0.3797 | 0.3487 |
| M50 | 0.5779 | 3 | 2 | 0.4879 | 0.5065 | 0.3689 |
| M51 | 0.5833 | 6 | 3 | 0.5195 | 0.3462 | 0.4235 |
| M52 | 0.8176 | 3 | 2 | 0.2983 | 0.0135 | 0.2538 |
| M53 | 0.3851 | 5 | 3 | 0.6599 | 0.4730 | 0.5856 |
| 平均Mean | 0.6593 | 3.3019 | 2.2264 | 0.4304 | 0.3480 | 0.3489 |
Table 4 Polymorphism information of 53 InDel markers
| 标记名称Marker name | 主等位基因频率Major allele frquency | 基因型数Genotype number | Na 等位基因数 Number of allele | Hs 基因多样性指数 Gene diversity index | He 期望杂合度 Expected heterozygosity | PIC 多态性信息含量 Polymorphic information content |
|---|---|---|---|---|---|---|
| M1 | 0.8101 | 3 | 2 | 0.3076 | 0.3038 | 0.2603 |
| M2 | 0.8734 | 3 | 2 | 0.2211 | 0.1013 | 0.1967 |
| M3 | 0.7278 | 3 | 2 | 0.3962 | 0.1646 | 0.3177 |
| M4 | 0.8354 | 2 | 2 | 0.2750 | 0.3291 | 0.2372 |
| M5 | 0.6519 | 3 | 2 | 0.4539 | 0.443 | 0.3509 |
| M6 | 0.6582 | 3 | 2 | 0.4499 | 0.5063 | 0.3487 |
| M7 | 0.6282 | 3 | 2 | 0.4671 | 0.3333 | 0.3580 |
| M8 | 0.7025 | 3 | 2 | 0.4180 | 0.5190 | 0.3306 |
| M9 | 0.4808 | 6 | 3 | 0.6335 | 0.5513 | 0.5620 |
| M10 | 0.4286 | 3 | 3 | 0.6208 | 0 | 0.5404 |
| M11 | 0.5321 | 3 | 2 | 0.4979 | 0.5769 | 0.3740 |
| M12 | 0.6329 | 3 | 2 | 0.4647 | 0.5316 | 0.3567 |
| M13 | 0.5506 | 3 | 2 | 0.4949 | 0.7215 | 0.3724 |
| M14 | 0.5886 | 3 | 2 | 0.4843 | 0.6456 | 0.3670 |
| M15 | 0.6042 | 3 | 2 | 0.4783 | 0.2639 | 0.3639 |
| M16 | 0.4464 | 6 | 3 | 0.6435 | 0.2679 | 0.5697 |
| M17 | 0.3667 | 8 | 4 | 0.7212 | 0.4889 | 0.6703 |
| M18 | 0.5506 | 3 | 2 | 0.4949 | 0.2405 | 0.3724 |
| M19 | 0.5000 | 3 | 2 | 0.5000 | 0.3333 | 0.3750 |
| M20 | 0.8671 | 2 | 2 | 0.2305 | 0.2658 | 0.2039 |
| M21 | 0.8101 | 2 | 2 | 0.3076 | 0.3797 | 0.2603 |
| M22 | 0.5513 | 3 | 2 | 0.4947 | 0.2564 | 0.3724 |
| M23 | 0.9679 | 3 | 2 | 0.0620 | 0.0385 | 0.0601 |
| M24 | 0.6456 | 5 | 4 | 0.5042 | 0.5696 | 0.4375 |
| M25 | 0.5064 | 3 | 2 | 0.4999 | 0.8077 | 0.3750 |
| M26 | 0.5949 | 2 | 2 | 0.4820 | 0.8101 | 0.3658 |
| M27 | 0.5506 | 3 | 2 | 0.4949 | 0.5696 | 0.3724 |
| M28 | 0.5972 | 3 | 2 | 0.4811 | 0.4167 | 0.3654 |
| M29 | 0.7500 | 3 | 2 | 0.3750 | 0.2917 | 0.3047 |
| M30 | 0.7278 | 3 | 2 | 0.3962 | 0.2911 | 0.3177 |
| M31 | 0.5789 | 3 | 2 | 0.4875 | 0.1579 | 0.3687 |
| M32 | 0.6753 | 3 | 2 | 0.4385 | 0.2078 | 0.3424 |
| M33 | 0.7756 | 3 | 2 | 0.3480 | 0.3974 | 0.2875 |
| M34 | 0.6731 | 3 | 2 | 0.4401 | 0.2949 | 0.3432 |
| M35 | 0.4333 | 4 | 3 | 0.6516 | 0.0222 | 0.5784 |
| M36 | 0.8924 | 3 | 2 | 0.1920 | 0.1139 | 0.1736 |
| M37 | 0.9026 | 3 | 2 | 0.1758 | 0.1429 | 0.1604 |
| M38 | 0.6899 | 4 | 3 | 0.4423 | 0.3671 | 0.3644 |
| M39 | 0.8103 | 3 | 2 | 0.3074 | 0.1034 | 0.2601 |
| M40 | 0.8291 | 3 | 2 | 0.2834 | 0.3165 | 0.2432 |
| M41 | 0.6218 | 3 | 2 | 0.4703 | 0.5513 | 0.3597 |
| M42 | 0.6866 | 4 | 3 | 0.4740 | 0.0896 | 0.4216 |
| M43 | 0.6014 | 3 | 2 | 0.4794 | 0.1014 | 0.3645 |
| M44 | 0.7722 | 3 | 2 | 0.3519 | 0.3797 | 0.2900 |
| M45 | 0.8734 | 3 | 2 | 0.2211 | 0.2025 | 0.1967 |
| M46 | 0.7975 | 3 | 2 | 0.3230 | 0.3797 | 0.2709 |
| M47 | 0.6392 | 2 | 2 | 0.4612 | 0.7215 | 0.3549 |
| M48 | 0.5294 | 3 | 2 | 0.4983 | 0.1569 | 0.3741 |
| M49 | 0.6582 | 3 | 2 | 0.4499 | 0.3797 | 0.3487 |
| M50 | 0.5779 | 3 | 2 | 0.4879 | 0.5065 | 0.3689 |
| M51 | 0.5833 | 6 | 3 | 0.5195 | 0.3462 | 0.4235 |
| M52 | 0.8176 | 3 | 2 | 0.2983 | 0.0135 | 0.2538 |
| M53 | 0.3851 | 5 | 3 | 0.6599 | 0.4730 | 0.5856 |
| 平均Mean | 0.6593 | 3.3019 | 2.2264 | 0.4304 | 0.3480 | 0.3489 |
| 编号 Code | 分子身份证 Molecular ID | 编号 Code | 分子身份证 Molecular ID | 编号 Code | 分子身份证 Molecular ID | 编号 Code | 分子身份证 Molecular ID |
|---|---|---|---|---|---|---|---|
| GD01 | 0A40012232122 | GD21 | 0A20811132122 | GZ02 | 1111121111221 | HB02 | 1327812242222 |
| GD02 | 3610811121152 | GD22 | 3514411121122 | GZ03 | 2222212142331 | ZJ01 | 8814421121142 |
| GD03 | 5A69822122112 | GD23 | 7653412332322 | GZ04 | 3222312112321 | ZJ02 | 6912731151151 |
| GD04 | 6A14811531252 | GD24 | 2514411121242 | GZ05 | 5223213212231 | ZJ03 | B528811224156 |
| GD05 | 8A20011231252 | GD25 | 4A14811531202 | GZ06 | 5223212112331 | FJ01 | 3529811264156 |
| GD06 | 1A14811131152 | GD26 | 4340721122201 | GZ07 | 3223212112301 | FJ02 | 2A02221723112 |
| GD07 | 2261211332242 | GD27 | 6513211132102 | GZ08 | 4331211122102 | FJ03 | 6630215142302 |
| GD08 | 0940012232222 | GX01 | 8A20811252451 | GZ09 | 1411412222121 | SD01 | 4640321153102 |
| GD09 | 7A50812322352 | GX02 | 3660811142252 | GZ10 | 3713312225256 | SD02 | 3212514123102 |
| GD10 | 7A50012322352 | GX03 | 0650312142341 | GZ11 | 3681011205305 | HN01 | 9940717153102 |
| GD11 | C690611321262 | GX04 | 6A40312242344 | GZ12 | BA93011125305 | XJ01 | 3660351143102 |
| GD12 | 8A24611321152 | GX05 | 3650811152131 | GZ13 | BA33312725303 | SX01 | 4330312153402 |
| GD13 | 2A14611321112 | GX06 | 6370811242141 | SC01 | 4220312112001 | LN01 | 6770725131202 |
| GD14 | AA20811623122 | GX07 | 0619811422152 | SC02 | 1230211132121 | LN02 | 4900326130102 |
| GD15 | 9970811131152 | GX08 | 4635825242354 | SC03 | 9320312142331 | JL01 | 3AA2311126506 |
| GD16 | 7660812232152 | GX09 | 6615812222151 | SC04 | B617221424255 | JL02 | 36A2321226506 |
| GD17 | 1A10812132212 | GX10 | 4619812422112 | CQ01 | 4320315142301 | HLJ01 | 3AA0372126606 |
| GD18 | 4A10812232222 | GX11 | 8630962541252 | CQ02 | 8620312142331 | HLJ02 | 3AA2321126506 |
| GD19 | 6A14811531222 | GX12 | 6615812242111 | YN01 | 4344612123102 | XL01 | 2223121121242 |
| GD20 | 8B52311231152 | GZ01 | 1111121111211 | HB01 | 6325715242222 |
Table 5 Molecular ID code of 79 plum germplasms
| 编号 Code | 分子身份证 Molecular ID | 编号 Code | 分子身份证 Molecular ID | 编号 Code | 分子身份证 Molecular ID | 编号 Code | 分子身份证 Molecular ID |
|---|---|---|---|---|---|---|---|
| GD01 | 0A40012232122 | GD21 | 0A20811132122 | GZ02 | 1111121111221 | HB02 | 1327812242222 |
| GD02 | 3610811121152 | GD22 | 3514411121122 | GZ03 | 2222212142331 | ZJ01 | 8814421121142 |
| GD03 | 5A69822122112 | GD23 | 7653412332322 | GZ04 | 3222312112321 | ZJ02 | 6912731151151 |
| GD04 | 6A14811531252 | GD24 | 2514411121242 | GZ05 | 5223213212231 | ZJ03 | B528811224156 |
| GD05 | 8A20011231252 | GD25 | 4A14811531202 | GZ06 | 5223212112331 | FJ01 | 3529811264156 |
| GD06 | 1A14811131152 | GD26 | 4340721122201 | GZ07 | 3223212112301 | FJ02 | 2A02221723112 |
| GD07 | 2261211332242 | GD27 | 6513211132102 | GZ08 | 4331211122102 | FJ03 | 6630215142302 |
| GD08 | 0940012232222 | GX01 | 8A20811252451 | GZ09 | 1411412222121 | SD01 | 4640321153102 |
| GD09 | 7A50812322352 | GX02 | 3660811142252 | GZ10 | 3713312225256 | SD02 | 3212514123102 |
| GD10 | 7A50012322352 | GX03 | 0650312142341 | GZ11 | 3681011205305 | HN01 | 9940717153102 |
| GD11 | C690611321262 | GX04 | 6A40312242344 | GZ12 | BA93011125305 | XJ01 | 3660351143102 |
| GD12 | 8A24611321152 | GX05 | 3650811152131 | GZ13 | BA33312725303 | SX01 | 4330312153402 |
| GD13 | 2A14611321112 | GX06 | 6370811242141 | SC01 | 4220312112001 | LN01 | 6770725131202 |
| GD14 | AA20811623122 | GX07 | 0619811422152 | SC02 | 1230211132121 | LN02 | 4900326130102 |
| GD15 | 9970811131152 | GX08 | 4635825242354 | SC03 | 9320312142331 | JL01 | 3AA2311126506 |
| GD16 | 7660812232152 | GX09 | 6615812222151 | SC04 | B617221424255 | JL02 | 36A2321226506 |
| GD17 | 1A10812132212 | GX10 | 4619812422112 | CQ01 | 4320315142301 | HLJ01 | 3AA0372126606 |
| GD18 | 4A10812232222 | GX11 | 8630962541252 | CQ02 | 8620312142331 | HLJ02 | 3AA2321126506 |
| GD19 | 6A14811531222 | GX12 | 6615812242111 | YN01 | 4344612123102 | XL01 | 2223121121242 |
| GD20 | 8B52311231152 | GZ01 | 1111121111211 | HB01 | 6325715242222 |
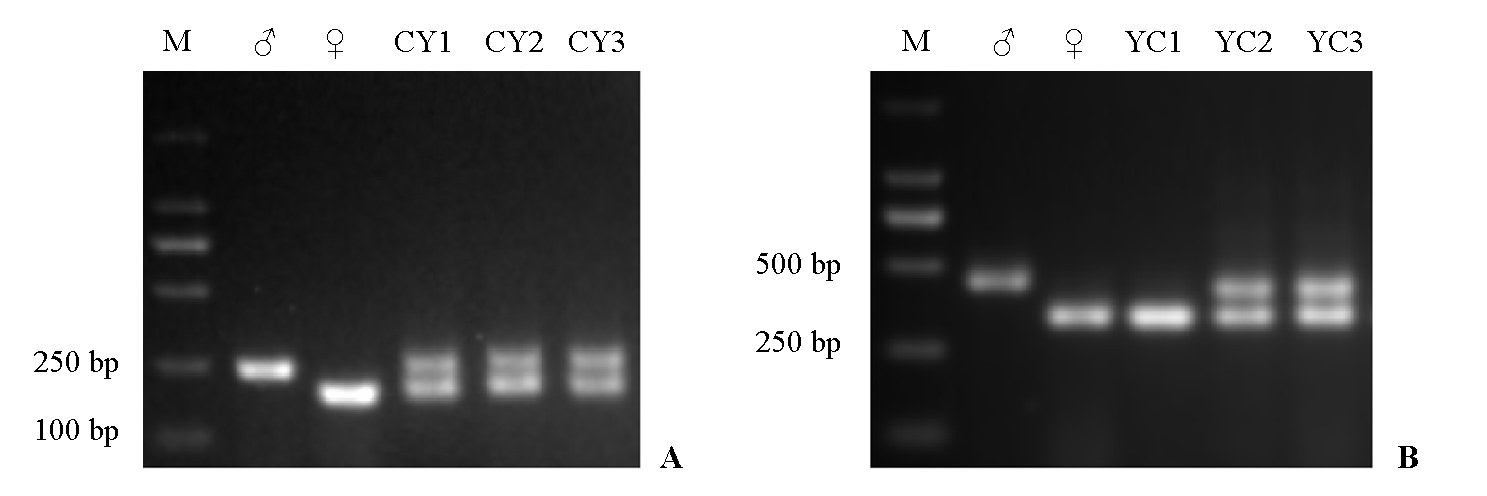
Fig. 3 Identification of hybrid seedling progeny using primers M12(A)and M44(B) A:♂:‘Yunkai 1’;♀:‘Congzao 1’;CY:‘Congzao 1’בYunkai 1’F1. B:♂:‘Congzao 1’;♀:‘Yunkai 1’;YC:‘Yunkai 1’בCongzao 1’F1. M:Marker 2000.
| [1] |
pmid: 6247908 |
| [2] |
|
|
陈红, 杨迤然. 2014. 贵州李资源遗传多样性及亲缘关系的ISSR分析. 果树学报, 31 (2):175-180.
|
|
| [3] |
doi: 10.3389/fpls.2018.00021 pmid: 29515596 |
| [4] |
|
| [5] |
|
| [6] |
|
|
郭翠红, 何业华, 冯筠庭, 陈程杰, 林文秋, 栾爱萍, 张雅芬. 2015. 广东省李产业发展现状调查. 经济林研究, 33 (1):141-146.
|
|
| [7] |
|
|
何业华, 潘建君, 杨向晖, 刘朝阳, 李楚豪, 夏靖娴, 张伟, 彭伟卓, 郝静静, 徐荣波, 孔文辉, 钟乙中. 2023. 李新品种‘从早1号早李’. 园艺学报, 50 (12):2765-2766.
doi: 10.16420/j.issn.0513-353x.2021-0317 |
|
| [8] |
doi: 10.16420/j.issn.0513-353x.2020-0067 |
|
何业华, 杨向晖, 栾爱萍, 刘成明, 胡桂兵, 林顺权, 秦永华, 夏靖娴, 傅嘉欣, 赵杰堂, 高用顺, 张志珂, 温瑞明, 陈世凯, 罗学优, 池琼云, 卢仕威. 2020. 华南李新品种‘云开1号’. 园艺学报, 47 (S2):2891-2892.
|
|
| [9] |
doi: 10.3864/j.issn.0578-1752.2016.12.002 |
|
胡振帮, 高运来, 齐照明, 蒋洪蔚, 刘春燕, 辛大伟, 胡国华, 潘校成, 陈庆山. 2016. 作物分子身份证构建软件ID analysis的编制. 中国农业科学, 49 (12):2255-2266.
|
|
| [10] |
doi: 10.16420/j.issn.0513-353x.2019-0982 |
|
林存学, 杨晓华, 刘海荣. 2020. 东北寒地96份李种质资源表型性状遗传多样性分析. 园艺学报, 47 (10):1917-1929.
|
|
| [11] |
|
|
刘均. 2017. 基于SRAP和IRAP标记的诱变李嫁接株系遗传变异分析[硕士论文]. 成都: 四川农业大学.
|
|
| [12] |
|
| [13] |
|
|
刘栓桃, 张志刚, 王荣花, 王立华, 李巧云, 赵智中. 2019. 基于InDel标记的大白菜育种材料分子身份证构建. 中国蔬菜,(2):34-41.
|
|
| [14] |
|
|
刘威生, 章秋平, 马小雪, 张玉萍, 刘家成, 张玉君, 刘硕, 刘宁, 徐铭. 2019. 新中国果树科学研究70年——李. 果树学报, 36 (10):1320-1338.
|
|
| [15] |
doi: 10.16420/j.issn.0513-353x.2016-0519 |
|
刘源霞, 兰进好, 柏素花, 孙晓红, 刘春晓, 张玉刚, 戴洪义. 2017. 苹果抗炭疽菌叶枯病基因SNP和InDel标记的HRM筛选. 园艺学报, 44 (2):215-222.
doi: 10.16420/j.issn.0513-353x.2016-0519 |
|
| [16] |
|
| [17] |
|
| [18] |
doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z pmid: 10655034 |
| [19] |
|
|
宋海岩, 孙淑霞, 李靖, 涂美艳, 王玲利, 徐子鸿, 陈栋, 江国良. 2023. 基于SSR标记检测与重测序技术的3个李品种鉴定与遗传背景简析. 中国南方果树, 52 (3):94-101.
|
|
| [20] |
doi: 10.16420/j.issn.0513-353x.2021-0935 |
|
汤雨晴, 杨惠栋, 闫承璞, 王斯妤, 王雨亭, 胡钟东, 朱方红. 2023. 基于重测序的‘金兰柚’基因组InDel标记的开发及应用. 园艺学报, 50 (1):15-26.
doi: 10.16420/j.issn.0513-353x.2021-0935 |
|
| [21] |
|
|
王珏, 王燕, 张静, 陈涛, 王磊, 陈清, 汤浩茹, 王小蓉. 2020. 中国樱桃InDel标记开发及其在蔷薇科果树中通用性评价. 园艺学报, 47 (1):98-110.
doi: 10.16420/j.issn.0513-353x.2019-0062 |
|
| [22] |
doi: 10.3864/j.issn.0578-1752.2019.03.017 |
|
魏潇, 章秋平, 刘宁, 张玉萍, 徐铭, 刘硕, 张玉君, 马小雪, 刘威生. 2019. 不同来源中国李(Prunus salicina L.) 的多样性与近缘种关系. 中国农业科学, 52 (3):568-578.
doi: 10.3864/j.issn.0578-1752.2019.03.017 |
|
| [23] |
|
|
徐溢岑. 2022. 李属植物资源叶绿体基因组序列分析与分子鉴定[硕士论文]. 重庆: 西南大学.
|
|
| [24] |
|
|
张加延, 周恩. 1998. 中国果树志 · 李卷. 北京: 中国林业出版社.
|
|
| [25] |
|
|
张淑文, 俞浙萍, 孙鹂, 梁森苗, 郑锡良, 颜丽菊, 黄颖宏, 戚行江. 2024. 杨梅种质资源遗传多样性分析与DNA指纹图谱构建. 分子植物育种, 22 (5):1488-1500.
|
|
| [26] |
doi: 10.16420/j.issn.0513-353x.2014-0679 |
|
左力辉, 韩志校, 梁海永, 杨敏生. 2015. 不同产地中国李资源遗传多样性SSR分析. 园艺学报, 42 (1):111-118.
doi: 10.16420/j.issn.0513-353x.2014-0679 |
| [1] | QIN Zilu, XU Zhengkang, DAI Xiaogang, CHEN Yingnan. Genetic Diversity Analysis and Core Collection Construction of Magnolia biondii Germplasm [J]. Acta Horticulturae Sinica, 2024, 51(8): 1823-1832. |
| [2] | YUAN Na, XU Qinyuan, XU Zhaolong, ZHOU Ling, LIU Xiaoqing, CHEN Xin, DU Jianchang. The Development and Verification of SNP Lquid Chips for Common Bean Based on Targeted Sequencing Technology [J]. Acta Horticulturae Sinica, 2024, 51(5): 1017-1032. |
| [3] | YOU Ping, YANG Jin, ZHOU Jun, HUANG Aijun, BAO Minli, and YI Long, . Genetic Diversity Analysis of Candidatus Liberibacter Asiaticus Based on Different Types of Prophage [J]. Acta Horticulturae Sinica, 2024, 51(4): 727-736. |
| [4] | YOU Yuanyuan, WANG Shuai, FANG Yingying, QI Cheng, LI Shupei, ZHANG Ying, CHEN Yuhui, LIU Wei, LIU Fuzhong, SHU Jinshuai. Variation Characteristics of Insertion-Deletion(InDel)and Molecular Markers Development and Application in Eggplant Based on Whole Genome Re-sequencing Data [J]. Acta Horticulturae Sinica, 2024, 51(3): 520-532. |
| [5] | YUE Linqing, WU Yichao, HE Xingxing, XIANG Mingyan, WANG Jin, WU Zheng, ZHU Shiguo, DING Mengqi, ZHANG Jian, FANG Xiaomei, YI Zelin. SSR Marker Association Analysis and Fingerprint Construction of Main Traits of Plum Fruit [J]. Acta Horticulturae Sinica, 2024, 51(11): 2495-2509. |
| [6] | WANG Rongbo, WANG Haiyun, ZHANG Qianrong, CAI Songling, LI Benjin, ZHANG Meixiang, LIU Peiqing. Genetic Diversity of Ralstonia solanacearum Strains Isolated from Fujian Province and Evaluation of Tomato Rootstocks for Resistance to Bacterial Wilt [J]. Acta Horticulturae Sinica, 2024, 51(11): 2540-2554. |
| [7] | YAO Yuan, DENG Lijun, HU Juan, TANG Xiaoyu, WANG Tie, LI Hang, SUN Guochao, XIONG Bo, LIAO Ling, and WANG Zhihui, . Whole Genome Resequencing Analysis of‘Cuihongli’Plum and Its Early-Ripening Bud Sport Mutation [J]. Acta Horticulturae Sinica, 2024, 51(10): 2255-2266. |
| [8] | LI Chunniu, SU Qun, LI Xianmin, HUANG Zhanwen, SUN Mingyan, LU Jiashi, WANG Hongyan, and BU Zhaoyang. SSR Molecular Markers Development and Parentage Relationship Identification Based on Whole Genomic Sequences of Jasminum sambac [J]. Acta Horticulturae Sinica, 2024, 51(10): 2343-2357. |
| [9] | GUAN Xiayu, CHEN Xihong, GUO Jing, LÜ Haoyang, LIANG Chenyuan, GAO Fangluan. Genetic Variability and Host Adaption of Cymbidium Mosaic Virus [J]. Acta Horticulturae Sinica, 2024, 51(1): 203-212. |
| [10] | YANG Xinyu , GOU Yunxia , WU Wanying , HE Runlin , DING Qiong , and WANG Dongliang, . A New Plum Blossom‘Wanjianglve’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 173-174. |
| [11] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, WANG Jun. Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [12] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, YAO Xingwei. Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [13] | DUAN Kaihang, WANG Xiaoling, MAO Yongmin, WANG Yao, REN Yongxiang, REN Liuliu, SHEN Lianying. Analysis of Genetic Diversity of Wild Jujube Germplasm Resources Based on Quantitative Characters [J]. Acta Horticulturae Sinica, 2023, 50(12): 2568-2576. |
| [14] | LIU Chenglang, FENG Di, CAO Zonghong, YAN Suyun, ZHANG Yongqing, XU Xiangzeng, GAO Shide, YE Junli, CHAI Lijun, XIE Zongzhou, DENG Xiuxin. Comparative Analysis of Genetic Background of‘Dongshi Zaoyou’,‘Shatianyou’and‘Shuijingyou’Pummelos [J]. Acta Horticulturae Sinica, 2023, 50(11): 2337-2349. |
| [15] | XIE Qian, ZHANG Shiyan, JIANG Lai, DING Mingyue, LIU Lingling, WU Rujian, CHEN Qingxi. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application [J]. Acta Horticulturae Sinica, 2023, 50(11): 2350-2364. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd