
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (6): 1215-1229.doi: 10.16420/j.issn.0513-353x.2022-0339
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
YANG Mengxia, LIU Xiaolin, CAO Xue, WEI Kai, NING Yu, YANG Pei, LI Shanshan, CHEN Ziyue, WANG Xiaoxuan, GUO Yanmei, DU Yongchen, LI Junming, LIU Lei, LI Xin, HUANG Zejun( )
)
Received:2022-11-29
Revised:2023-02-27
Online:2023-06-25
Published:2023-06-27
Contact:
* (E-mail:CLC Number:
YANG Mengxia, LIU Xiaolin, CAO Xue, WEI Kai, NING Yu, YANG Pei, LI Shanshan, CHEN Ziyue, WANG Xiaoxuan, GUO Yanmei, DU Yongchen, LI Junming, LIU Lei, LI Xin, HUANG Zejun. Construction and Application of a CRISPR/Cas9 System for Multiplex Gene Editing in Tomato[J]. Acta Horticulturae Sinica, 2023, 50(6): 1215-1229.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0339
| 引物Primer | 引物序列(5′-3′)Primer sequence |
|---|---|
| MGET-17 | TTCATCTCGGGTCTCGTTTGATCCACGATGATCTCCGTTGTTTTAGAGCTAGAAATAG |
| MGET-18 | TTCATCTCGGGTCTCtAAACTCTGCCTCTCTTGATGCCCTGACCGTTGATAGTGGATAG |
| MGET-19 | TTCATCTCGGGTCTCGCTTGTCCATTGCCACATTAGTGGGTTTTAGAGCTAGAAATAG |
| MGET-20 | TTCATCTCGGGTCTCtAAACCCCAACTCAAGCCACAAAGTGACATATCCTACTGGTACG |
| MGET-21 | TTCATCTCGGGTCTCGTTTGGTGAAGAACCTGCTATTCAGTTTTAGAGCTAGAAATAG |
| MGET-43 | TTCATCTCGGGTCTCtAAACATTAGTAAACATCATGCCACAAACACACATGCTAATATTC |
| MGET-23 | TTCATCTCGGGTCTCGTTTGCTTTGTGGCTTGAGTTGGGGTTTTAGAGCTAGAAATAG |
| MGET-24 | TTCATCTCGGGTCTCtAAACTGAATAGCAGGTTCTTCACTGACCGTTGATAGTGGATAG |
| MGET-44 | TTCATCTCGGGTCTCGCTTGTGGCATGATGTTTACTAATGTTTTAGAGCTAGAAATAG |
| MGET-26 | TTCATCTCGGGTCTCtAAACAACGGAGATCATCGTGGATTGACATATCCTACTGGTACG |
| MGET-27 | TTCATCTCGGGTCTCGTTTGGGGCATCAAGAGAGGCAGAGTTTTAGAGCTAGAAATAG |
| MGET-28 | TTCATCTCGGGTCTCtAAACCCACTAATGTGGCAATGGACAAACACACATGCTAATATTC |
| ZP121 | GAACAAAAGCCTTGAGAACC |
| ZP122 | GTCGAAATCCCACACATCAC |
| ZP123 | CAACTAACCTTATCACTGAATCC |
| ZP124 | TGGTTAACTTTCCAGGGAGC |
| ZP125 | GATGCTGGCATTTGTGTGATTC |
| ZP126 | ACGACTAGAACCGTCAACGA |
| ZP129 | CAAAGAGTGACATGCAATGC |
| ZP130 | CCTCTTGAGAAGTAATGTTCC |
| ZP149 | GCAGAGACAAAAAGAATGGG |
| ZP150 | CATGATTAATCAAAGGAGGTTC |
| ZP191 | GCCGAACACATCAACATTTC |
| ZP192 | CCACACGTCTCGTCAGAATG |
| Cas9-909F | GCAGCTCTCCAAGGACACAT |
| Cas9-2450R | CGTGAGTTCTTCTGGCCCTT |
Table 1 Sequence information of primers
| 引物Primer | 引物序列(5′-3′)Primer sequence |
|---|---|
| MGET-17 | TTCATCTCGGGTCTCGTTTGATCCACGATGATCTCCGTTGTTTTAGAGCTAGAAATAG |
| MGET-18 | TTCATCTCGGGTCTCtAAACTCTGCCTCTCTTGATGCCCTGACCGTTGATAGTGGATAG |
| MGET-19 | TTCATCTCGGGTCTCGCTTGTCCATTGCCACATTAGTGGGTTTTAGAGCTAGAAATAG |
| MGET-20 | TTCATCTCGGGTCTCtAAACCCCAACTCAAGCCACAAAGTGACATATCCTACTGGTACG |
| MGET-21 | TTCATCTCGGGTCTCGTTTGGTGAAGAACCTGCTATTCAGTTTTAGAGCTAGAAATAG |
| MGET-43 | TTCATCTCGGGTCTCtAAACATTAGTAAACATCATGCCACAAACACACATGCTAATATTC |
| MGET-23 | TTCATCTCGGGTCTCGTTTGCTTTGTGGCTTGAGTTGGGGTTTTAGAGCTAGAAATAG |
| MGET-24 | TTCATCTCGGGTCTCtAAACTGAATAGCAGGTTCTTCACTGACCGTTGATAGTGGATAG |
| MGET-44 | TTCATCTCGGGTCTCGCTTGTGGCATGATGTTTACTAATGTTTTAGAGCTAGAAATAG |
| MGET-26 | TTCATCTCGGGTCTCtAAACAACGGAGATCATCGTGGATTGACATATCCTACTGGTACG |
| MGET-27 | TTCATCTCGGGTCTCGTTTGGGGCATCAAGAGAGGCAGAGTTTTAGAGCTAGAAATAG |
| MGET-28 | TTCATCTCGGGTCTCtAAACCCACTAATGTGGCAATGGACAAACACACATGCTAATATTC |
| ZP121 | GAACAAAAGCCTTGAGAACC |
| ZP122 | GTCGAAATCCCACACATCAC |
| ZP123 | CAACTAACCTTATCACTGAATCC |
| ZP124 | TGGTTAACTTTCCAGGGAGC |
| ZP125 | GATGCTGGCATTTGTGTGATTC |
| ZP126 | ACGACTAGAACCGTCAACGA |
| ZP129 | CAAAGAGTGACATGCAATGC |
| ZP130 | CCTCTTGAGAAGTAATGTTCC |
| ZP149 | GCAGAGACAAAAAGAATGGG |
| ZP150 | CATGATTAATCAAAGGAGGTTC |
| ZP191 | GCCGAACACATCAACATTTC |
| ZP192 | CCACACGTCTCGTCAGAATG |
| Cas9-909F | GCAGCTCTCCAAGGACACAT |
| Cas9-2450R | CGTGAGTTCTTCTGGCCCTT |
| 基因名称 Gene name | 基因ID Gene ID | 靶点序列(5′-3′) Target sequence | PAM区 PAM area |
|---|---|---|---|
| Green flesh(GF) | Solyc08g080090 | CCACTAATGTGGCAATGGA | CCT |
| Ovate(O) | Solyc02g085500 | ATCCACGATGATCTCCGTT | GGG |
| Locule number(LC) | Solyc02g083950 | ATTAGTAAACATCATGCCA | CCA |
| Tangerine(T) | Solyc10g081650 | CCCAACTCAAGCCACAAAG | CCC |
| Uniform(U) | Solyc10g008160 | GTGAAGAACCTGCTATTCA | TGG |
| SlMYB12(Y) | Solyc01g079620 | GGGCATCAAGAGAGGCAGA | TGG |
Table 2 The target nucleotide sequence of multiple genes
| 基因名称 Gene name | 基因ID Gene ID | 靶点序列(5′-3′) Target sequence | PAM区 PAM area |
|---|---|---|---|
| Green flesh(GF) | Solyc08g080090 | CCACTAATGTGGCAATGGA | CCT |
| Ovate(O) | Solyc02g085500 | ATCCACGATGATCTCCGTT | GGG |
| Locule number(LC) | Solyc02g083950 | ATTAGTAAACATCATGCCA | CCA |
| Tangerine(T) | Solyc10g081650 | CCCAACTCAAGCCACAAAG | CCC |
| Uniform(U) | Solyc10g008160 | GTGAAGAACCTGCTATTCA | TGG |
| SlMYB12(Y) | Solyc01g079620 | GGGCATCAAGAGAGGCAGA | TGG |
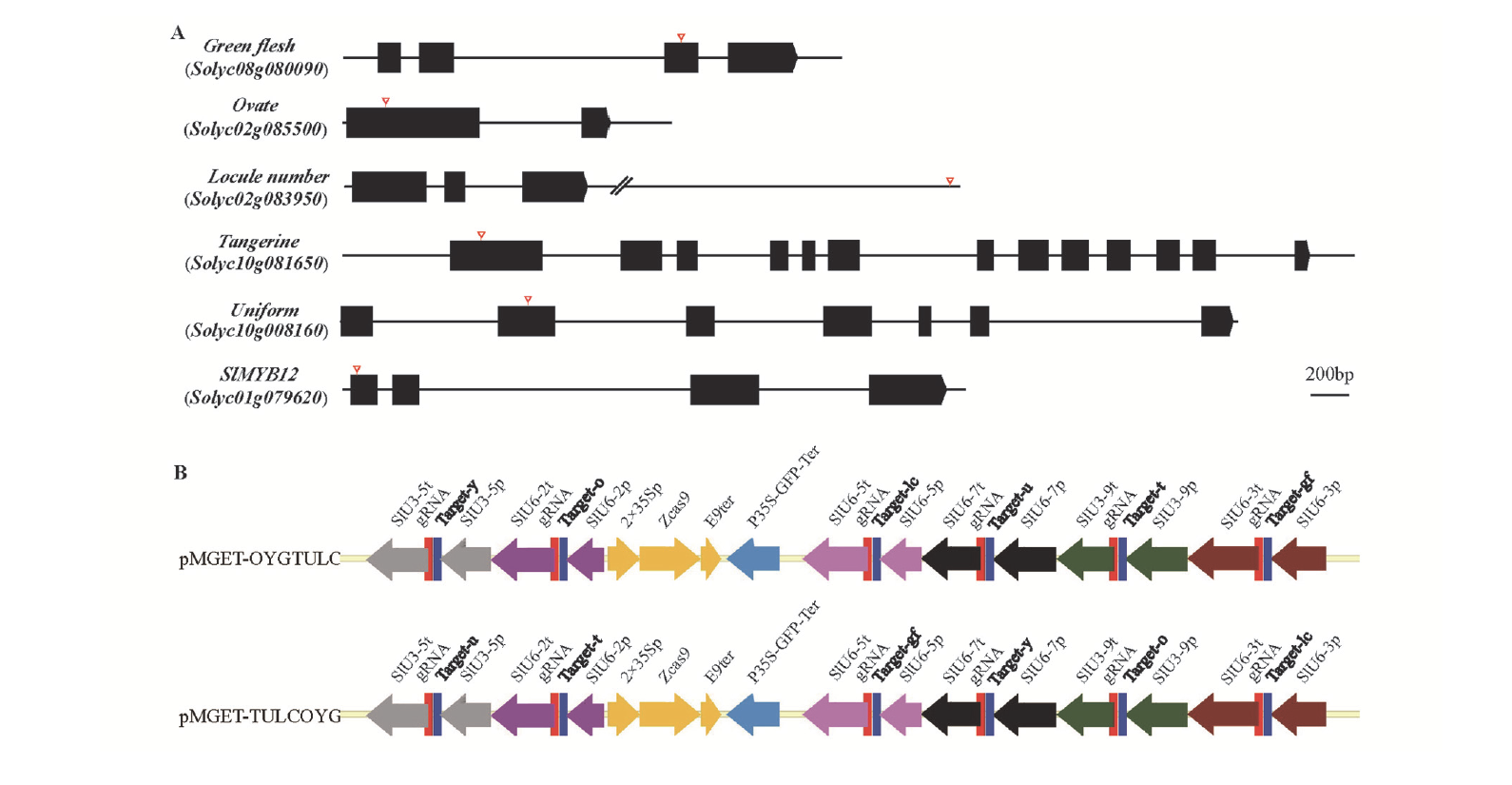
Fig. 3 Diagram of gene target location and gene editing vector structure A:Represents intron-exon structure of GF,O,LC,T,U and Y gene and the location of their target sites,and the red triangle indicates the target site;B:Represents the vector diagram of sgRNA driven by different promoters.
| 载体名称 Vector name | 基因名称 Gene name | 启动子 Promoter | 测序的毛状根数 Number of roots for sequencing | 单基因编辑 Single gene editing | 6个基因同时编辑 Six genes are edited at the same time | ||
|---|---|---|---|---|---|---|---|
| 数量Number | 效率/% Efficiency | 数量Number | 效率/% Efficiency | ||||
| pMGET- | Green flesh | SlU6-3p | 50 | 45 | 83.33 | ||
| OYGTULC | Tangerine | SlU3-9p | 50 | 41 | 75.93 | ||
| Uniform | SlU6-7p | 50 | 39 | 72.22 | |||
| SlMYB12 | SlU3-5p | 50 | 45 | 83.33 | |||
| Ovate | SlU6-2p | 50 | 29 | 53.70 | |||
| Locule number | SlU6-5p | 50 | 45 | 83.33 | |||
| 小计 Subtotal | 22 | 44.00 | |||||
| pMGET- | Green flesh | SlU6-5p | 51 | 49 | 96.08 | ||
| TULCOYG | Tangerine | SlU6-2p | 51 | 50 | 98.04 | ||
| Uniform | SlU3-5p | 51 | 10 | 19.61 | |||
| SlMYB12 | SlU6-7p | 51 | 49 | 96.08 | |||
| Ovate | SlU3-9p | 51 | 38 | 74.51 | |||
| Locule number | SlU6-3p | 51 | 50 | 98.04 | |||
| 小计 Subtotal | 6 | 11.76 | |||||
Table 3 Editing efficiency of different genes of two vectors
| 载体名称 Vector name | 基因名称 Gene name | 启动子 Promoter | 测序的毛状根数 Number of roots for sequencing | 单基因编辑 Single gene editing | 6个基因同时编辑 Six genes are edited at the same time | ||
|---|---|---|---|---|---|---|---|
| 数量Number | 效率/% Efficiency | 数量Number | 效率/% Efficiency | ||||
| pMGET- | Green flesh | SlU6-3p | 50 | 45 | 83.33 | ||
| OYGTULC | Tangerine | SlU3-9p | 50 | 41 | 75.93 | ||
| Uniform | SlU6-7p | 50 | 39 | 72.22 | |||
| SlMYB12 | SlU3-5p | 50 | 45 | 83.33 | |||
| Ovate | SlU6-2p | 50 | 29 | 53.70 | |||
| Locule number | SlU6-5p | 50 | 45 | 83.33 | |||
| 小计 Subtotal | 22 | 44.00 | |||||
| pMGET- | Green flesh | SlU6-5p | 51 | 49 | 96.08 | ||
| TULCOYG | Tangerine | SlU6-2p | 51 | 50 | 98.04 | ||
| Uniform | SlU3-5p | 51 | 10 | 19.61 | |||
| SlMYB12 | SlU6-7p | 51 | 49 | 96.08 | |||
| Ovate | SlU3-9p | 51 | 38 | 74.51 | |||
| Locule number | SlU6-3p | 51 | 50 | 98.04 | |||
| 小计 Subtotal | 6 | 11.76 | |||||
| [1] |
Barry C S, McQuinn R P, Chung M Y, Besuden A, Giovannoni J J. 2008. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol, 147 (1):179-187.
doi: 10.1104/pp.108.118430 pmid: 18359841 |
| [2] |
Brooks C, Nekrasov V, Lippman Z B, van Eck J. 2014. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 system. Plant Physiol, 166 (3):1292-1297.
doi: 10.1104/pp.114.247577 URL |
| [3] |
Čermák T, Curtin S J, Gil-Humanes J, Čegan R, Kono T J Y, Konečná E, Belanto J J, Starker C G, Mathre J W, Greenstein R L, Voytas D F. 2017. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29 (6):1196-1217.
doi: 10.1105/tpc.16.00922 URL |
| [4] | Chaudhary J, Alisha A, Bhatt V, Chandanshive S, Kumar N, Mir Z, Kumar A, Yadav S K, Shivaraj S M, Sonah H, Deshmukh R. 2019. Mutation breeding in tomato:Advances,applicability and challenges. Plants(Basel), 8 (5):128. |
| [5] |
Chen K, Gao C. 2014. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Reports, 33 (4):575-583.
doi: 10.1007/s00299-013-1539-6 pmid: 24277082 |
| [6] | Cui Xia, Zhang Shuai-bin. 2017. The utilization and prospect of genome editing in horticultural crops. Acta Horticulturae Sinica, 44 (9):1787-1795. (in Chinese) |
| 崔霞, 张率斌. 2017. 基因编辑技术及其在园艺作物中的应用和展望. 园艺学报, 44 (9):1787-1795. | |
| [7] |
Deng L, Wang H, Sun C, Li Q, Jiang H, Du M, Li C B, Li C. 2018. Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J Genet Genomics, 45 (1):51-54.
doi: S1673-8527(17)30178-9 pmid: 29157799 |
| [8] | Du H Y, Zeng X R, Zhao M, Cui X P, Wang Q, Yang H, Cheng H, Yu D Y. 2016. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. Biotechnol, 217:90-97. |
| [9] |
Du M, Zhou K, Liu Y, Deng L, Zhang X, Lin L, Zhou M, Zhao W, Wen C, Xing J, Li C B, Li C. 2020. A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J, 102 (5):1090-1100.
doi: 10.1111/tpj.v102.5 URL |
| [10] |
Gao Y, Zhu N, Zhu X, Wu M, Jiang C Z, Grierson D, Luo Y, Shen W, Zhong S, Fu D Q, Qu G. 2019. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas 9 CNR and NOR mutants. Hortic Res, 6:39.
doi: 10.1038/s41438-019-0122-x |
| [11] |
Hisamatsu S, Sakaue M, Takizawa A, Kato T, Kamoshita M, Ito J, Kashiwazaki N. 2015. Knockout of targeted gene in porcine somatic cells using zinc-finger nuclease. Anim Sci J, 86 (2):132-137.
doi: 10.1111/asj.12259 pmid: 25187232 |
| [12] |
Isaacson T, Ronen G, Zamir D, Hirschberg J. 2002. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell, 14 (2):333-342.
doi: 10.1105/tpc.010303 pmid: 11884678 |
| [13] |
Kimura S, Sinha N. 2008. Tomato(Solanum lycopersicum):a model fruit-bearing crop. CSH Protocols, DOI:10.1101/pdb.emo105.
doi: 10.1101/pdb.emo105 |
| [14] |
Kiss T, Solymosy F. 1990. Molecular analysis of a U3 RNA gene locus in tomato:transcription signals,the coding region,expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res, 18 (8):1941-1949.
pmid: 2336383 |
| [15] |
Klap C, Yeshayahou E, Bolger A M, Arazi T, Gupta S K, Shabtai S, Usadel B, Salts Y, Barg R. 2017. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J, 15 (5):634-647.
doi: 10.1111/pbi.2017.15.issue-5 URL |
| [16] |
Kwon C T, Heo J, Lemmon Z H, Capua Y, Hutton S F, van Eck J, Park S J, Lippman Z B. 2020. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat Biotechnol, 38 (2):182-188.
doi: 10.1038/s41587-019-0361-2 |
| [17] |
Li R, Li R, Li X, Fu D, Zhu B, Tian H, Luo Y, Zhu H. 2018a. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol J, 16 (2):415-427.
doi: 10.1111/pbi.2018.16.issue-2 URL |
| [18] |
Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C. 2018b. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol, 36:1160-1163.
doi: 10.1038/nbt.4273 URL |
| [19] |
Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y, Zhu H. 2018c. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci, 9:559.
doi: 10.3389/fpls.2018.00559 URL |
| [20] |
Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, Huang Z, Li J, Zhang C, Wang T, Zhang Y, Wang A, Zhang Y, Lin K, Li C, Xiong G, Xue Y, Mazzucato A, Causse M, Fei Z, Giovannoni J J, Chetelat R T, Zamir D, Städler T, Li J, Ye Z, Du Y, Huang S. 2014. Genomic analyses provide insights into the history of tomato breeding. Nat Genet, 46 (11):1220-1226.
doi: 10.1038/ng.3117 pmid: 25305757 |
| [21] | Liu Chun-xia, Gui Hua-ping, Wen Qin, Liu Yu-bo, Gao Yang, Zhang Xiu-juan, Cui Rui-jie, Xu Jian-ping. 2020. CRISPR/Cas 9 genome editing system optimization in tomato. Molecular Plant Breeding, 18 (20):6716-6724. (in Chinese) |
| 刘春霞, 桂花萍, 文琴, 刘宇博, 高阳, 张秀娟, 崔瑞洁, 许建平. 2020. 番茄CRISPR/Cas9基因编辑效率的优化. 分子植物育种, 18 (20):6716-6724. | |
| [22] |
Liu J, van Eck J, Cong B, Tanksley S D. 2002. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci U S A, 99 (20):13302-13306.
doi: 10.1073/pnas.162485999 URL |
| [23] |
Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, Lu Y, Wang Q, Li C, Zhai Q. 2019. MYC 2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell, 31 (1):106-127.
doi: 10.1105/tpc.18.00405 URL |
| [24] |
Lu J, Pan C, Li X, Huang Z, Shu J, Wang X, Lu X, Pan F, Hu J, Zhang H, Su W, Zhang M, Du Y, Liu L, Guo Y, Li J. 2021. OBV(obscure vein),a C2H 2 zinc finger transcription factor,positively regulates chloroplast development and bundle sheath extension formation in tomato(Solanum lycopersicum)leaf veins. Hortic Res, 8 (1):230.
doi: 10.1038/s41438-021-00659-z |
| [25] |
Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G. 2015. A robust CRISPR/Cas 9 system for convenient,high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant, 8 (8):1274-1284.
doi: 10.1016/j.molp.2015.04.007 URL |
| [26] |
Montecillo J A V, Chu L L, Bae H. 2020. CRISPR-Cas 9 system for plant genome editing:current approaches and emerging developments. Agronomy, 10 (7):1033.
doi: 10.3390/agronomy10071033 URL |
| [27] |
Muños S, Ranc N, Botton E, Bérard A, Rolland S, Duffé P, Carretero Y, Le Paslier M C, Delalande C, Bouzayen M, Brunel D, Causse M. 2011. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol, 156 (4):2244-2254.
doi: 10.1104/pp.111.173997 pmid: 21673133 |
| [28] |
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. 2017. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep, 7 (1):482.
doi: 10.1038/s41598-017-00578-x pmid: 28352080 |
| [29] |
Nonaka S, Arai C, Takayama M, Matsukura C, Ezura H. 2017. Efficient increase of ɣ-aminobutyric acid(GABA)content in tomato fruits by targeted mutagenesis. Sci Rep, 7 (1):7057.
doi: 10.1038/s41598-017-06400-y |
| [30] |
Powell A L, Nguyen C V, Hill T, Cheng K L, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, Lopez-Baltazar J, Barry C S, Liu Y, Chetelat R, Granell A, van Deynze A, Giovannoni J J, Bennett A B. 2012. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science, 336 (6089):1711-1715.
doi: 10.1126/science.1222218 pmid: 22745430 |
| [31] |
Rodríguez-Leal D, Lemmon Z H, Man J, Bartlett M E, Lippman Z B. 2017. Engineering quantitative trait variation for crop improvement by genome editing. Cell, 171 (2):470-480.
doi: S0092-8674(17)30988-1 pmid: 28919077 |
| [32] |
Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso M A, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, Federici F, Sinha N, Deal R B, Bailey-Serres J, Brady S M. 2014. Hairy root transformation using Agrobacterium rhizo genes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol, 166 (2):455-469.
doi: 10.1104/pp.114.239392 |
| [33] |
Shan Qi-wei, Gao Cai-xia. 2015. Research progress of genome editing and derivative technologies in plants. Hereditas(Beijing), 37 (10):953-973. (in Chinese)
doi: 10.16288/j.yczz.15-156 pmid: 26496748 |
| 单奇伟, 高彩霞. 2015. 植物基因组编辑及衍生技术最新研究进展. 遗传, 37 (10):953-973. | |
| [34] |
Sun N, Zhao H. 2013. Transcription activator-like effector nucleases(TALENs):A highly efficient and versatile tool for genome editing. Biotechnol Bioeng, 110 (7):1811-1821.
doi: 10.1002/bit.24890 URL |
| [35] |
Sun X J, Hu Z, Chen R, Jiang Q Y, Song G H, Zhang H, Xi Y J. 2015. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep, 5:10342.
doi: 10.1038/srep10342 pmid: 26022141 |
| [36] | van der Knaap E, Chakrabarti M, Chu Y H, Clevenger J P, Illa-Berenguer E, Huang Z, Keyhaninejad N, Mu Q, Sun L, Wang Y, Wu S. 2014. What lies beyond the eye:the molecular mechanisms regulating tomato fruit weight and shape. Front Plant Sci, 5:227. |
| [37] |
Waibel F, Filipowicz W. 1990. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res, 18 (12):3451-3458.
pmid: 2362802 |
| [38] |
Wang C, Shen L, Fu Y, Yan C, Wang K. 2015. A simple CRISPR/Cas 9 system for multiplex genome editing in rice. J Genet Genomics, 42 (12):703-706.
doi: 10.1016/j.jgg.2015.09.011 URL |
| [39] | Wang Dan, Wang Mi, Liu Jun, Zhou Xiaohui, Liu Songyu, Yang Yan, Zhuang Yong. 2022. Cloning of U6 promoters and establishment of CRISPR/Cas 9 mediated gene editing system in eggplant. Acta Horticulturae Sinica, 49 (4):791-800. (in Chinese) |
|
王丹, 王谧, 刘军, 周晓慧, 刘松瑜, 杨艳, 庄勇. 2022. 茄子U6启动子克隆及CRISPR/Cas9介导的基因编辑体系建立. 园艺学报, 49 (4):791-800.
doi: 10.16420/j.issn.0513-353x.2020-0265 URL |
|
| [40] | Wang Juan, Wang Baike, Li Ning, Yang Tao, Patiguli ASMUTOLA, Yu Qinghui. 2020. Application progress of gene editing technology in tomato breeding. Plant Physiology Journal, 56 (12):2606-2616. (in Chinese) |
| 王娟, 王柏柯, 李宁, 杨涛, 帕提古丽 · 艾斯木托拉, 余庆辉. 2020. 基因编辑技术在番茄育种中的应用进展. 植物生理学报, 56 (12):2606-2616. | |
| [41] | Wang Mu-jia, Zhu Jian-kang. 2018. Simplification of multiplex gene editing systems in plants. Chinese Bulletin of Life Sciences, 30 (9):980-986. (in Chinese) |
| 王木桂, 朱健康. 2018. 化繁为简——植物多基因编辑体系的优化. 生命科学, 30 (9):980-986. | |
| [42] |
Xing H L, Dong L, Wang Z P, Zhang H Y, Han C Y, Liu B, Wang X C, Chen Q J. 2014. A CRISPR/Cas 9 toolkit for multiplex genome editing in plants. BMC Plant Biol, 14:327.
doi: 10.1186/s12870-014-0327-y URL |
| [43] | Yang Meng-xia. 2020. Creation of tomato male sterile material by gene editing[M. D. Dissertation]. Beijing: Chinese Academy of Agricultural Sciences. (in Chinese) |
| 杨孟霞. 2020. 利用基因编辑技术创制番茄雄性不育材料的研究[硕士论文]. 北京: 中国农业科学院. | |
| [44] |
Ye J, Wang X, Hu T, Zhang F, Wang B, Li C, Yang T, Li H, Lu Y, Giovannoni J J, Zhang Y, Ye Z. 2017. An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell, 29 (9):2249-2268.
doi: 10.1105/tpc.17.00211 URL |
| [45] |
Yu W, Wang L, Zhao R, Sheng J, Zhang S, Li R, Shen L. 2019. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol, 19 (1):354.
doi: 10.1186/s12870-019-1939-z |
| [46] |
Zhang Aiping, Liu Jiangna, Yan Jianjun, Zhang Xiying, Bai Yunfeng. 2022. Progress and prospect of genome editing in tomato. Acta Horticulturae Sinica, 49 (1):221-232. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0976 URL |
|
张爱萍, 刘江娜, 闫建俊, 张西英, 白云凤. 2022. 番茄基因编辑研究进展和前景. 园艺学报, 49 (1):221-232.
doi: 10.16420/j.issn.0513-353x.2020-0976 URL |
|
| [47] |
Zhang Z, Mao Y, Ha S, Liu W, Botella J R, Zhu J K. 2016. A multiplex CRISPR/Cas 9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep, 35 (7):1519-1533.
doi: 10.1007/s00299-015-1900-z URL |
| [48] |
Zhou Xiang-chun, Xing Yong-zhong. 2016. The application of genome editing in identification of plant gene function and crop breeding. Hereditas(Beijing), 38 (3):227-242. (in Chinese)
doi: 10.16288/j.yczz.15-327 pmid: 27001477 |
| 周想春, 邢永忠. 2016. 基因组编辑技术在植物基因功能鉴定及作物育种中的应用. 遗传, 38 (3):227-242. | |
| [49] |
Zsögön A, Čermák T, Naves E R, Notini M M, Edel K H, Weinl S, Freschi L, Voytas D F, Kudla J, Peres L E P. 2018. De novo domestication of wild tomato using genome editing. Nat Biotechnol, 36 (12):1211-1216.
doi: 10.1038/nbt.4272 |
| [1] | SU Yinling, YANG Zixiang, DAN Zhong, MA Jixian, YANG Long, LI Yirong, TANG Zhengfu, WANG Lingmin, and MU Wanfu. A New Tomato Cultivar‘Yun Tomato 68’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 63-64. |
| [2] | FAN Juan, SHEN Songzhen, MIAO Qingqing, ZHANG Lehui, YAO Enpeng, and PEI Zhuoqiang . A New Tomato Cultivar‘Weihong 16’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 65-66. |
| [3] | ZHANG Hui, ZHU Weimin, ZHU Longying, YANG Xuedong, ZHANG Yingying, TIAN Shoubo, WAN Yanhui, LIU Yahui, YANG Zhijie. A New Cherry Tomato Cultivar‘Huying 9’ [J]. Acta Horticulturae Sinica, 2023, 50(6): 1379-1380. |
| [4] | LI Guobin, CAI Liangyu, XIAO Licheng, WANG Jiafa, ZHANG Dedi, ZHANG Junhong. Creating Pink Fruit and Fruit Without Green Shoulder Using CRISPR/ Cas9 Technology in Tomato [J]. Acta Horticulturae Sinica, 2023, 50(5): 985-999. |
| [5] | XU Yue, LI Zixiong, CHEN Jie, SUN Liang. Transcriptional Bases of the Regulation of 2,4-D on Tomato Fruit Shape [J]. Acta Horticulturae Sinica, 2023, 50(4): 802-814. |
| [6] | ZHENG Jinrong, NIE Jun, LI Yanhong, TAN Delong, XIE Yuming, ZHANG Changyuan. A Cherry Tomato Cultivar‘Yuekeda 101’ [J]. Acta Horticulturae Sinica, 2023, 50(4): 909-910. |
| [7] | LIU Yuhan, TAO Ning, WANG Qingguo, LI Qingqing. ABC Transporter SlABCG23 Regulates Jasmonic Acid Signaling Pathway in Tomato [J]. Acta Horticulturae Sinica, 2023, 50(3): 559-568. |
| [8] | SHI Hongli, LI La, GUO Cuimei, YU Tingting, JIAN Wei, YANG Xingyong. Isolation,Identification and Analysis of Biocontrol Ability of Biocontrol Strain TL1 Against Tomato Botrytis cinerea [J]. Acta Horticulturae Sinica, 2023, 50(1): 79-90. |
| [9] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [10] | ZHENG Jirong, WANG Tonglin, and HU Songshen. A New Tomato Cultivar‘Hangza 603’with High Quality [J]. Acta Horticulturae Sinica, 2022, 49(S2): 103-104. |
| [11] | ZHENG Jirong and WANG Tonglin. A New Tomato Cultivar‘Hangza 601’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 105-106. |
| [12] | ZHENG Jirong and WANG Tonglin. A New Cherry Tomato Cultivar‘Hangza 503’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 107-108. |
| [13] | HUANG Tingting, LIU Shuqin, ZHANG Yongzhi, LI Ping, ZHANG Zhihuan, and SONG Libo. A New Cherry Tomato Cultivar‘Yingshahong 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 109-110. |
| [14] | ZHANG Qianrong, LI Dazhong, QIU Boyin, LIN Hui, MA Huifei, YE Xinru, LIU Jianting, ZHU Haisheng, and WEN Qingfang. A New Tomato Cultivar‘Minnongke 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 73-74. |
| [15] | HAN Shuai, WU Jie, ZHANG Heqing, XI Yadong. Identification and Sequence Analysis of Tomato Spotted Wilt Orthotospovirus Infecting Lettuce in Sichuan [J]. Acta Horticulturae Sinica, 2022, 49(9): 2007-2016. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd