
园艺学报 ›› 2025, Vol. 52 ›› Issue (10): 2625-2641.doi: 10.16420/j.issn.0513-353x.2024-0833
边拴灵1, 邓昌蓉1, 樊露泽2, 吴延寻1, 任延靖1,3,*( )
)
收稿日期:2025-03-18
修回日期:2025-06-16
出版日期:2025-10-25
发布日期:2025-10-28
通讯作者:
基金资助:
BIAN Shuanling1, DENG Changrong1, FAN Luze2, WU Yanxun1, REN Yanjing1,3,*( )
)
Received:2025-03-18
Revised:2025-06-16
Published:2025-10-25
Online:2025-10-28
摘要:
以黄色芜菁W25和白色芜菁W21的不同组织为材料,构建芜菁cDNA酵母文库,库容1.5 × 107 CFU,插入片段平均长度 > 1 000 bp。通过构建的诱饵载体pGBKT7-PSY与构建的cDNA文库杂交,共获得了18个阳性互作蛋白,通过酵母双杂交及双分子荧光互补(BiFC)验证,明确了候选蛋白CDPK8与PSY存在互作关系。对CDPK家族进行鉴定,共鉴定出50个含有STKc_CAMK protein kinase和2个EF-hand结构域的CDPK基因(命名为BrrCDPK1 ~ BrrCDPK50),结构分析显示CDPK编码氨基酸数在411 ~ 660个之间,平均等电点为8.79,各CDPK蛋白之间保守基序数和结构差异不明显,均含有motif 1、motif 2、motif 3和motif 6,染色体定位显示BrrCDPKs在7条染色体上的分布是非随机的;系统发育分析表明芜菁BrrCDPK与拟南芥AtCDPK被聚为不同类,暗示其在进化过程中的基因复制具有一定的种属特异性,基因表达谱分析鉴定出14个在黄色芜菁中高表达的CDPK基因。
边拴灵, 邓昌蓉, 樊露泽, 吴延寻, 任延靖. 芜菁酵母文库的构建和PSY潜在互作蛋白BrrCDPK家族成员鉴定[J]. 园艺学报, 2025, 52(10): 2625-2641.
BIAN Shuanling, DENG Changrong, FAN Luze, WU Yanxun, REN Yanjing. Construction of Yeast Library and Identification of the BrrCDPK Gene Family of PSY Potential Interacting Proteins in Turnip[J]. Acta Horticulturae Sinica, 2025, 52(10): 2625-2641.
| 引物名称Primer name | 引物序列(5′-3′)Primer sequence |
|---|---|
| CDPK8-F | ATGGGAGGTTGTACCTCCA |
| CDPK8-R | TCACCGAGTTTTAGCTAAAGG |
| pGBKT7-CDPK8-F | TGGCCATGGAGGCCGAATTCATGGGAGGTTGTACCTCCA |
| pGBKT7-CDPK8-R | CGCTGCAGGTCGACGGATCCTCACCGAGTTTTAGCTAAAGG |
| pGADT7-CDPK8-F | CCATGGAGGCCAGTGAATTCATGGGAGGTTGTACCTCCA |
| pGADT7-CDPK8-R | AGCTCGAGCTCGATGGATCCTCACCGAGTTTTAGCTAAAGG |
| pGADT7-PSY-F | CCATGGAGGCCAGTGAATTCATGTCTTCTGTAGCAGTGTTA |
| pGADT7-PSY-R | AGCTCGAGCTCGATGGATCCTTAAGTTGTTCCTCTTGAACTT |
| G2-PSY-F | AGCTCGAGCTCGATGGATCCATGGGAGGTTGTACCTCCA |
| G2-PSY-R | CTACCCGGGAGCGGTACCCTCGAGTCACCGAGTTTTAGCTAAAGG |
| G4-CDPK8-F | AGCTCGAGCTCGATGGATCCATGTCTTCTGTAGCAGTGTTA |
| G4-CDPK8-R | GAGCTCCTACCCGGGAGCGGTACCTTAAGTTGTTCCTCTTGAACTT |
表1 PSY互作蛋白验证引物序列
Table 1 Primer sequence of valivation PSY interacting proteins
| 引物名称Primer name | 引物序列(5′-3′)Primer sequence |
|---|---|
| CDPK8-F | ATGGGAGGTTGTACCTCCA |
| CDPK8-R | TCACCGAGTTTTAGCTAAAGG |
| pGBKT7-CDPK8-F | TGGCCATGGAGGCCGAATTCATGGGAGGTTGTACCTCCA |
| pGBKT7-CDPK8-R | CGCTGCAGGTCGACGGATCCTCACCGAGTTTTAGCTAAAGG |
| pGADT7-CDPK8-F | CCATGGAGGCCAGTGAATTCATGGGAGGTTGTACCTCCA |
| pGADT7-CDPK8-R | AGCTCGAGCTCGATGGATCCTCACCGAGTTTTAGCTAAAGG |
| pGADT7-PSY-F | CCATGGAGGCCAGTGAATTCATGTCTTCTGTAGCAGTGTTA |
| pGADT7-PSY-R | AGCTCGAGCTCGATGGATCCTTAAGTTGTTCCTCTTGAACTT |
| G2-PSY-F | AGCTCGAGCTCGATGGATCCATGGGAGGTTGTACCTCCA |
| G2-PSY-R | CTACCCGGGAGCGGTACCCTCGAGTCACCGAGTTTTAGCTAAAGG |
| G4-CDPK8-F | AGCTCGAGCTCGATGGATCCATGTCTTCTGTAGCAGTGTTA |
| G4-CDPK8-R | GAGCTCCTACCCGGGAGCGGTACCTTAAGTTGTTCCTCTTGAACTT |
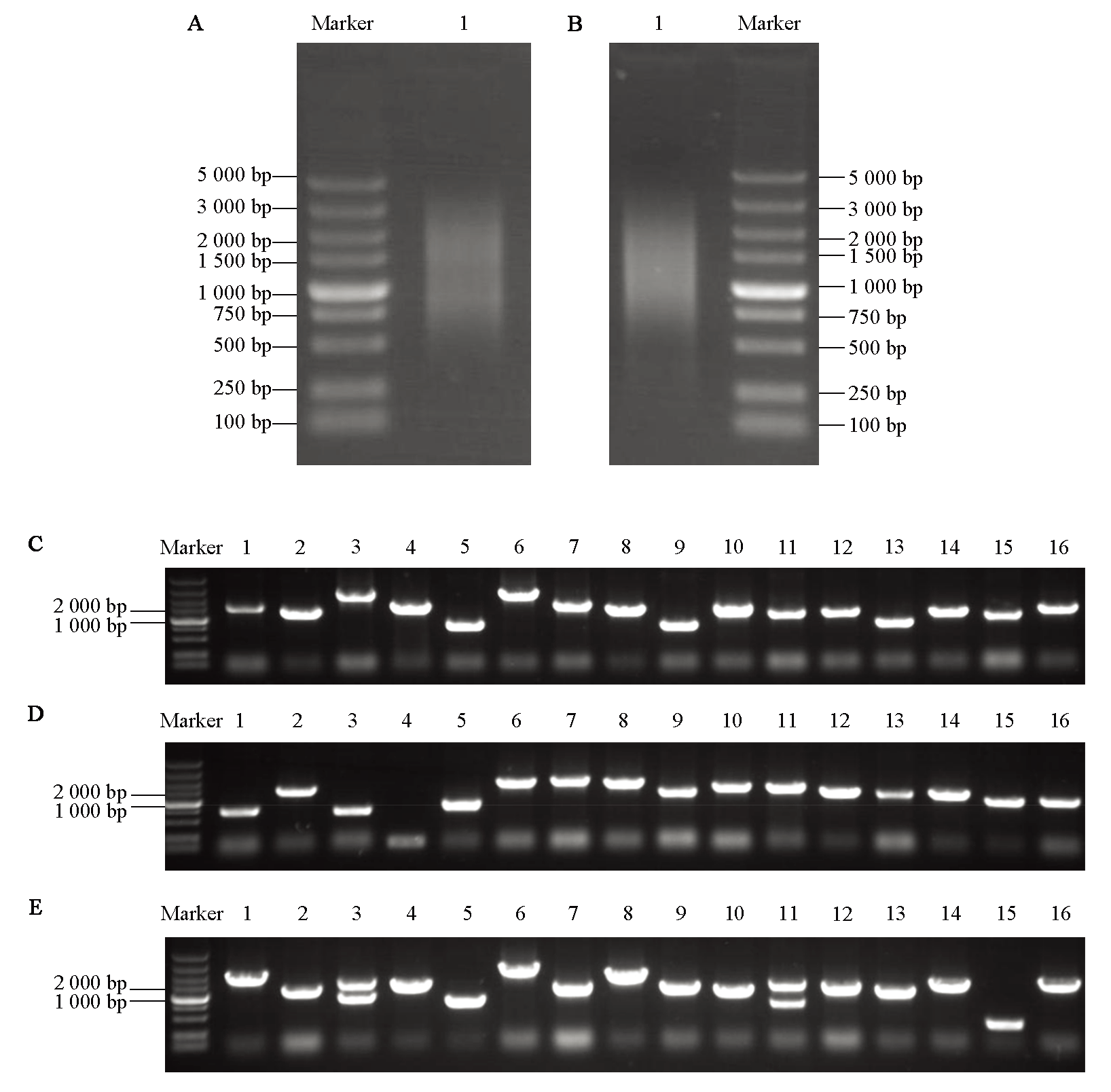
图1 芜菁酵母cDNA文库的构建及鉴定 A:cDNA合成电泳检测;B:cDNA均一化处理电泳检测;C:初级文库插入片段鉴定;D:次级文库插入片段鉴定;E:酵母文库插入片段鉴定。1 ~ 16:不同克隆的菌落PCR检测结果
Fig. 1 Construction and identification of yeast cDNA library in turnip A:Electrophoresis detection map of cDNA synthesis;B:Electrophoresis detection map of cDNA homogenization;C:Identification of inserts in the primary library;D:Identification of inserts in the secondary library;E:Identification of inserts in the yeast two-hybrid library. 1-16:Electrophoretic detection results of colony PCR with different clones

图3 芜菁PSY互作蛋白的筛选 A:1/100稀释悬浮液涂布DDO平板覆盖效果;B:1/1 000稀释悬浮液涂布DDO平板覆盖效果;C、D:芜菁PSY在SD/-Ade/-His/-Leu/-Trp/X/A(QDO/X/A)平板筛选的蓝色克隆平板正、反面照片;E:对QDO/X/A平板中能正常生长的菌落进行电泳检测。1 ~ 24:不同克隆的菌落PCR检测结果
Fig. 3 PSY interaction protein screening in turnip A:Covering effect of 1/100 dilution suspension on DDO plate;B:Covering effect of 1/1 000 dilution suspension on DDO plate;C,D:Front and back view of blue clonal plate screened by PSY on SD/-Ade/-His/-Leu/-Trp/X/A plates;E:Electrophoresis detection map of colonies that normally grow in QDO/X/A plates. 1-24:Electrophoretic detection results of colony PCR with different clones
| 序号No. | 样本名Sample name | 基因注释Gene annotation | 登录号GenBank | 基因ID Gene ID |
|---|---|---|---|---|
| C1 | 1237-BD-1-1 1237-BD-4 1237-BD-10 | 白菜低分子量热休克蛋白(BcHSP17.6)mRNA,完整CDS Brassica rapa low molecular weight heat-shock protein(BcHSP17.6)mRNA,complete CDS | AF022217.1 | BraA06g020450.3C |
| C2 | 1237-BD-2 1237-BD-6 1237-BD-18 | 白菜核前体mRNA的调节域含蛋白1A(LOC103855848),转录本变体X2 Brassica rapa regulation of nuclear pre-mRNA domain-containing protein 1A(LOC103855848),transcript variant X2 | XM_009132891.3 | BraA03g004120.3C |
| C3 | 1237-BD-3-2 | 白菜嘌呤渗透酶1(LOC103838976),mRNA Brassica rapa purine permease 1(LOC103838976) | XM_009115443.3 | BraA09g036510.3C |
| C4 | 1237-BD-8-1 | 白菜Golgi SNAP受体复合体成员1-2(LOC103858165),转录变体X2 Brassica rapa Golgi SNAP receptor complex member 1-2(LOC103858165),transcript variant X2 | XM_018657146.2 | BraA03g023640.3C |
| C5 | 1237-BD-9 | 白菜translin(LOC103857651),转录变体X1 Brassica rapa translin(LOC103857651),transcript variant X1 | XM_009134861.3 | BraA03g019260.3C |
| C6 | 1237-BD-11 | 白菜17.6 kDⅠ类热休克蛋白2-like(LOC103843285) Brassica rapa 17.6 kD classⅠheat shock protein 2-like (LOC103843285) | XM_009119989.3 | BraA05g014680.3C |
| C7 | 1237-BD-12 | 甘蓝型油菜多细胞器RNA编辑因子1,mitochondrial(LOC106437506) Brassica napus multiple organellar RNA editing factor 1,mitochondrial(LOC106437506) | XM_013878402.3 | BraA03g049050.3C |
| C8 | 1237-BD-13 | 白菜RNA聚合酶Ⅱ转录亚基26的推定介质(LOC103833180) Brassica rapa putative mediator of RNA polymeraseⅡtranscription subunit 26(LOC103833180) | XM_033282535.1 | BraA10g022490.3C |
| C9 | 1237-BD-14-2 | 甘蓝型油菜CDPK相关激酶2(LOC106362920),转录变体X2 Brassica napus CDPK-related kinase 2(LOC106362920),transcript variant X2 | XM_022695160.2 | BraA01g033890.3C |
| C10 | 1237-BD-15-1 | 甘蓝型油菜formyltetrahydrofolate deformylase1,线粒体(LOC106402097),转录变体X4 Brassica napus formyltetrahydrofolate deformylase 1,mitochondrial(LOC106402097),transcript variant X4 | XM_048781377.1 | BraA06g041130.3C |
| C11 | 1237-BD-15-2 | 白菜 patellin-4(LOC103840314),转录变体X2 Brassica rapa patellin-4(LOC103840314),transcript variant X2 | XM_009116810.2 | BraA09g034310.3C |
| C12 | 1237-BD-17-1 | 甘蓝型油菜硫氧还蛋白O2,线粒体(LOC106352370),转录变体X1 Brassica napus thioredoxin O2,mitochondrial(LOC106352370),transcript variant X1 | XM_048739086.1 | BraA09g033920.3C |
| C13 | 1237-BD-17-2 | 白菜同源箱亮氨酸拉链蛋白ATHB-6(LOC103837893) Brassica rapa homeobox-leucine zipper protein ATHB-6(LOC103837893) | XM_009114280.3 | BraA09g054870.3C |
| C14 | 1237-BD-20 | 白菜未鉴定的LOC103863008(LOC103863008),转录变体X3 Brassica rapa uncharacterized LOC103863008(LOC103863008),transcript variant X3 | XM_033289696.1 | BraA04g002940.3C |
| C15 | 1237-BD-21 | 白菜蛋白MET1,chloroplastic(LOC103829781) Brassica rapa protein MET1,chloroplastic(LOC103829781) | XM_009105478.3 | BraA05g016190.3C |
| C16 | 1237-BD-22 | 白菜转座酶蛋白At4g04430(LOC103847917),转录变体X2 Brassica rapa probable transposase-like protein At4g04430(LOC103847917),transcript variant X2 | XM_018655836.2 | 未检索到 Not retrieved |
| C17 | 1237-BD-23 | 甘蓝型油菜高迁移率B型蛋白2(LOC106416143),转录变体X2 Brassica napus high mobility group B protein 2(LOC106416143),transcript variant X2 | XM_048738078.1 | BraA08g027710.3C |
| C18 | 1237-BD-24 | 白菜E3泛素蛋白连接酶BRE1-like 1(LOC103866184) Brassica rapa E3 ubiquitin-protein ligase BRE1-like 1(LOC103866184) | XM_009144061.3 | BraA04g030860.3C |
表2 芜菁pGBKT7-PSY互作蛋白注释
Table 2 pGBKT7-PSY interaction protein annotation
| 序号No. | 样本名Sample name | 基因注释Gene annotation | 登录号GenBank | 基因ID Gene ID |
|---|---|---|---|---|
| C1 | 1237-BD-1-1 1237-BD-4 1237-BD-10 | 白菜低分子量热休克蛋白(BcHSP17.6)mRNA,完整CDS Brassica rapa low molecular weight heat-shock protein(BcHSP17.6)mRNA,complete CDS | AF022217.1 | BraA06g020450.3C |
| C2 | 1237-BD-2 1237-BD-6 1237-BD-18 | 白菜核前体mRNA的调节域含蛋白1A(LOC103855848),转录本变体X2 Brassica rapa regulation of nuclear pre-mRNA domain-containing protein 1A(LOC103855848),transcript variant X2 | XM_009132891.3 | BraA03g004120.3C |
| C3 | 1237-BD-3-2 | 白菜嘌呤渗透酶1(LOC103838976),mRNA Brassica rapa purine permease 1(LOC103838976) | XM_009115443.3 | BraA09g036510.3C |
| C4 | 1237-BD-8-1 | 白菜Golgi SNAP受体复合体成员1-2(LOC103858165),转录变体X2 Brassica rapa Golgi SNAP receptor complex member 1-2(LOC103858165),transcript variant X2 | XM_018657146.2 | BraA03g023640.3C |
| C5 | 1237-BD-9 | 白菜translin(LOC103857651),转录变体X1 Brassica rapa translin(LOC103857651),transcript variant X1 | XM_009134861.3 | BraA03g019260.3C |
| C6 | 1237-BD-11 | 白菜17.6 kDⅠ类热休克蛋白2-like(LOC103843285) Brassica rapa 17.6 kD classⅠheat shock protein 2-like (LOC103843285) | XM_009119989.3 | BraA05g014680.3C |
| C7 | 1237-BD-12 | 甘蓝型油菜多细胞器RNA编辑因子1,mitochondrial(LOC106437506) Brassica napus multiple organellar RNA editing factor 1,mitochondrial(LOC106437506) | XM_013878402.3 | BraA03g049050.3C |
| C8 | 1237-BD-13 | 白菜RNA聚合酶Ⅱ转录亚基26的推定介质(LOC103833180) Brassica rapa putative mediator of RNA polymeraseⅡtranscription subunit 26(LOC103833180) | XM_033282535.1 | BraA10g022490.3C |
| C9 | 1237-BD-14-2 | 甘蓝型油菜CDPK相关激酶2(LOC106362920),转录变体X2 Brassica napus CDPK-related kinase 2(LOC106362920),transcript variant X2 | XM_022695160.2 | BraA01g033890.3C |
| C10 | 1237-BD-15-1 | 甘蓝型油菜formyltetrahydrofolate deformylase1,线粒体(LOC106402097),转录变体X4 Brassica napus formyltetrahydrofolate deformylase 1,mitochondrial(LOC106402097),transcript variant X4 | XM_048781377.1 | BraA06g041130.3C |
| C11 | 1237-BD-15-2 | 白菜 patellin-4(LOC103840314),转录变体X2 Brassica rapa patellin-4(LOC103840314),transcript variant X2 | XM_009116810.2 | BraA09g034310.3C |
| C12 | 1237-BD-17-1 | 甘蓝型油菜硫氧还蛋白O2,线粒体(LOC106352370),转录变体X1 Brassica napus thioredoxin O2,mitochondrial(LOC106352370),transcript variant X1 | XM_048739086.1 | BraA09g033920.3C |
| C13 | 1237-BD-17-2 | 白菜同源箱亮氨酸拉链蛋白ATHB-6(LOC103837893) Brassica rapa homeobox-leucine zipper protein ATHB-6(LOC103837893) | XM_009114280.3 | BraA09g054870.3C |
| C14 | 1237-BD-20 | 白菜未鉴定的LOC103863008(LOC103863008),转录变体X3 Brassica rapa uncharacterized LOC103863008(LOC103863008),transcript variant X3 | XM_033289696.1 | BraA04g002940.3C |
| C15 | 1237-BD-21 | 白菜蛋白MET1,chloroplastic(LOC103829781) Brassica rapa protein MET1,chloroplastic(LOC103829781) | XM_009105478.3 | BraA05g016190.3C |
| C16 | 1237-BD-22 | 白菜转座酶蛋白At4g04430(LOC103847917),转录变体X2 Brassica rapa probable transposase-like protein At4g04430(LOC103847917),transcript variant X2 | XM_018655836.2 | 未检索到 Not retrieved |
| C17 | 1237-BD-23 | 甘蓝型油菜高迁移率B型蛋白2(LOC106416143),转录变体X2 Brassica napus high mobility group B protein 2(LOC106416143),transcript variant X2 | XM_048738078.1 | BraA08g027710.3C |
| C18 | 1237-BD-24 | 白菜E3泛素蛋白连接酶BRE1-like 1(LOC103866184) Brassica rapa E3 ubiquitin-protein ligase BRE1-like 1(LOC103866184) | XM_009144061.3 | BraA04g030860.3C |
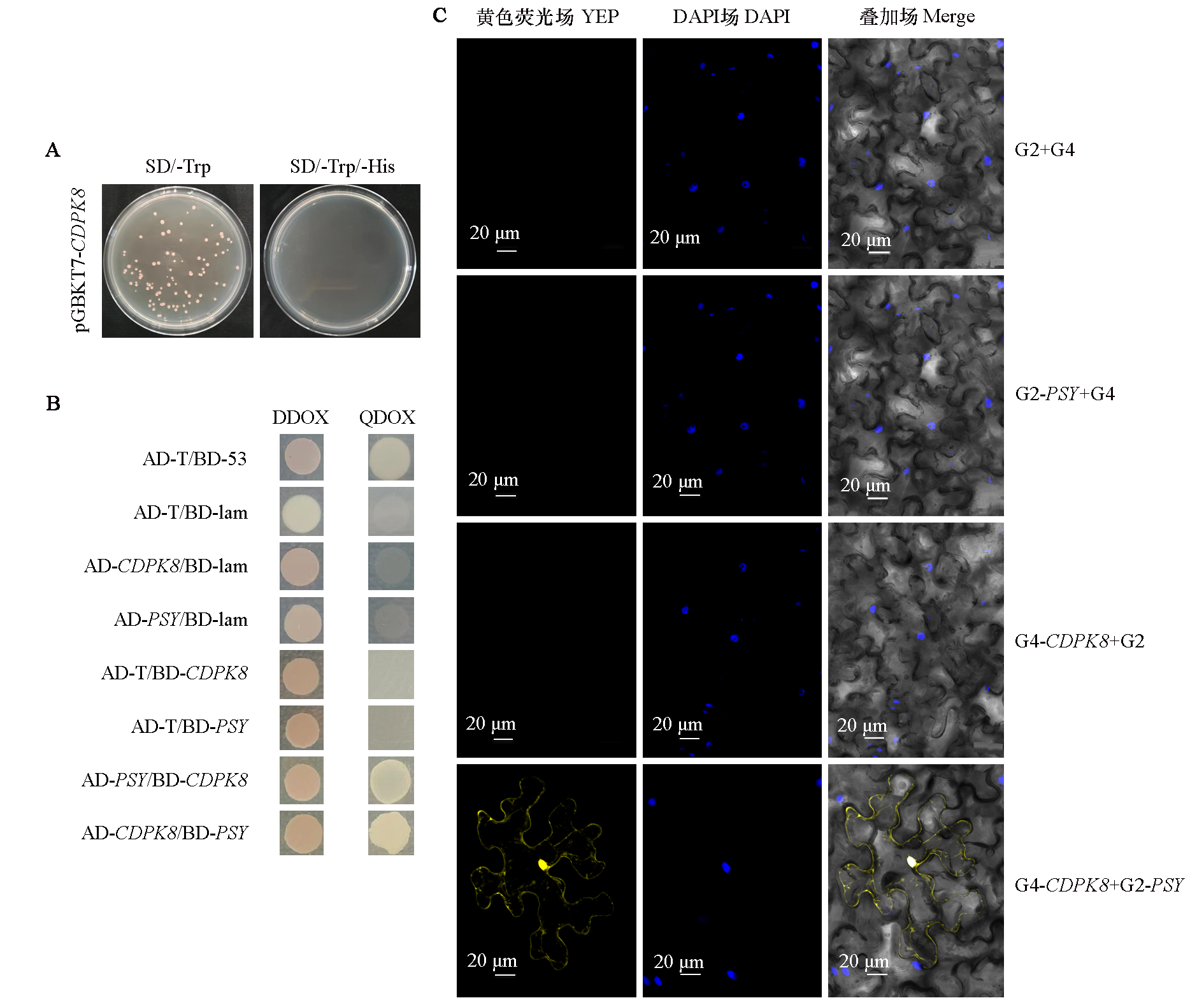
图4 芜菁PSY与互作蛋白CDPK点对点验证 A:pGBKT7-CDPK的诱饵载体的自激活鉴定;B:PSY与CDPK蛋白的酵母双杂交验证;C:PSY与CDPK在BiFC试验中的相互作用
Fig. 4 Verification of interaction relationship between PSY and CDPK protein in turnips A:pGBKT7-CDPK Y2H Gold strain self-activation detection;B:Verification of interaction relationship between PSY and CDPK protein by yeast two-hybrid;C:Interaction between PSY and CDPK in BiFC experiment
| 基因名 Gene name | 基因编号 Gene ID | 登录号 Accession number | cDNA长度/bp cDNA length | 氨基酸数 Number of amino acids | 分子量/kD Molecular weight | 等电点 Theoretical pI | 脂肪系数 Aliphatic index | 疏水性 GRAVY | |
|---|---|---|---|---|---|---|---|---|---|
| BrrCDPK1 | Cluster-30924.46777 | XM_009103718.3 | 1 737 | 578 | 64.58 | 8.95 | 83.39 | -0.317 | |
| BrrCDPK2 | Cluster-30924.137521 | XM_013807283.3 | 1 743 | 580 | 64.88 | 9.07 | 83.28 | -0.324 | |
| BrrCDPK3 | Cluster-30924.143914 | XM_013807283.3 | 1 731 | 576 | 64.47 | 9.01 | 83.68 | -0.318 | |
| BrrCDPK4 | Cluster-30924.13630 | XM_048738123.1 | 1 755 | 584 | 65.72 | 8.88 | 81.70 | -0.392 | |
| BrrCDPK5 | Cluster-30924.67984 | XM_010509132.2 | 1 806 | 601 | 67.46 | 8.99 | 88.80 | -0.318 | |
| BrrCDPK6 | Cluster-30924.158029 Cluster-30924.77034 Cluster-30924.77038 Cluster-30924.80376 | XM_009113894.3 | 1 785 | 594 | 66.74 | 9.06 | 83.43 | -0.374 | |
| BrrCDPK7 | Cluster-30924.58681 | XM_009113894.3 | 1 785 | 594 | 66.76 | 9.12 | 82.61 | -0.375 | |
| BrrCDPK8 | Cluster-30924.142390 | XM_009113894.3 | 1 785 | 594 | 66.77 | 9.06 | 83.60 | -0.373 | |
| BrrCDPK9 | Cluster-30924.157993 | XM_009113894.3 | 1 785 | 594 | 66.70 | 9.06 | 84.09 | -0.372 | |
| BrrCDPK10 | Cluster-30924.131630 | XM_013879035.3 | 1 704 | 567 | 63.43 | 8.84 | 86.84 | -0.218 | |
| BrrCDPK11 | Cluster-30924.136201 | XM_013792411.3 | 1 707 | 568 | 63.43 | 8.84 | 86.88 | -0.205 | |
| BrrCDPK12 | Cluster-30924.24510 | XM_013879035.3 | 1 704 | 567 | 63.59 | 8.67 | 85.47 | -0.219 | |
| BrrCDPK13 | Cluster-30924.57260 | XM_009140827.3 | 1 728 | 575 | 64.34 | 8.61 | 86.16 | -0.238 | |
| BrrCDPK14 | Cluster-30924.150034 | XM_013886734.3 | 1 713 | 570 | 63.80 | 8.85 | 86.74 | -0.251 | |
| BrrCDPK15 | Cluster-30924.150032 Cluster-30924.150033 | XM_009140827.3 | 1 728 | 575 | 64.38 | 8.70 | 86.16 | -0.243 | |
| BrrCDPK16 | Cluster-30924.14001 | XM_033289955.1 | 1 239 | 412 | 46.61 | 8.43 | 86.09 | -0.229 | |
| BrrCDPK17 | Cluster-30924.24231 | XM_013871943.3 | 1 236 | 411 | 46.25 | 8.00 | 88.42 | -0.203 | |
| BrrCDPK18 | Cluster-30924.82876 | XM_013895504.3 | 1 791 | 596 | 66.44 | 8.95 | 83.98 | -0.351 | |
| BrrCDPK19 | Cluster-30924.151434 | XM_013895504.3 | 1 791 | 596 | 66.45 | 8.88 | 84.80 | -0.343 | |
| BrrCDPK20 | Cluster-30924.142865 | XM_013895504.3 | 1 791 | 596 | 66.49 | 9.01 | 83.98 | -0.344 | |
| BrrCDPK21 | Cluster-30924.142866 | XM_009144190.3 | 1 791 | 596 | 66.45 | 8.95 | 83.98 | -0.356 | |
| BrrCDPK22 | Cluster-30924.146395 Cluster-30924.169091 | XM_013895504.3 | 1 791 | 596 | 66.44 | 8.88 | 83.98 | -0.350 | |
| BrrCDPK23 | Cluster-30924.52514 | XM_018622041.2 | 1 809 | 602 | 67.28 | 9.16 | 82.66 | -0.366 | |
| BrrCDPK24 | Cluster-30924.140928 Cluster-30924.155408 | XM_009144190.3 | 1 791 | 596 | 66.46 | 8.95 | 83.98 | -0.356 | |
| BrrCDPK25 | Cluster-30924.122791 | XM_013781701.1 | 1 734 | 577 | 64.40 | 8.52 | 84.85 | -0.260 | |
| BrrCDPK26 | Cluster-30924.24346 | XM_013892390.3 | 1 734 | 577 | 64.41 | 8.52 | 84.33 | -0.263 | |
| BrrCDPK27 | Cluster-30924.67985 | XM_009147578.3 | 1 983 | 660 | 74.20 | 9.02 | 85.41 | -0.248 | |
| BrrCDPK28 | Cluster-30924.67983 Cluster-30924.92828 | XM_009147578.3 | 1 983 | 660 | 74.31 | 9.07 | 86.15 | -0.246 | |
| BrrCDPK29 | Cluster-30924.11427 | XM_009147578.3 | 1 983 | 660 | 74.30 | 9.07 | 85.56 | -0.252 | |
| BrrCDPK30 | Cluster-30924.24345 | XM_009147578.3 | 1 983 | 660 | 74.33 | 9.07 | 84.97 | -0.254 | |
| BrrCDPK31 | Cluster-30924.77035 | XM_013732238.1 | 1 791 | 596 | 66.91 | 9.09 | 83.79 | -0.362 | |
| BrrCDPK32 | Cluster-30924.36432 | XM_013851068.3 | 1 791 | 596 | 66.95 | 9.13 | 83.79 | -0.355 | |
| BrrCDPK33 | Cluster-30924.77036 | XM_013893888.3 | 1 791 | 596 | 66.82 | 9.19 | 83.31 | -0.363 | |
| BrrCDPK34 | Cluster-30924.77037 | XM_013851068.3 | 1 791 | 596 | 66.91 | 9.13 | 83.79 | -0.362 | |
| BrrCDPK35 | Cluster-30924.114427 | XM_009147578.3 | 1 983 | 660 | 74.32 | 9.07 | 86.15 | -0.244 | |
| BrrCDPK36 | Cluster-69076.1 Cluster-69076.4 | XM_013786949.3 | 1 776 | 591 | 65.92 | 8.66 | 83.55 | -0.309 | |
| BrrCDPK37 | Cluster-69076.2 | XM_013786949.3 | 1 776 | 591 | 65.93 | 8.52 | 84.53 | -0.295 | |
| BrrCDPK38 | Cluster-69076.0 | XM_009151493.3 | 1 776 | 591 | 65.90 | 8.66 | 84.38 | -0.306 | |
| BrrCDPK39 | Cluster-30924.23018 | XM_013786949.3 | 1 776 | 591 | 65.95 | 8.77 | 83.89 | -0.314 | |
| BrrCDPK40 | Cluster-30924.12640 | XM_013787387.3 | 1 803 | 600 | 66.93 | 8.51 | 82.48 | -0.343 | |
| BrrCDPK41 | Cluster-30924.75342 | XM_048769751.1 | 1 242 | 413 | 47.04 | 6.68 | 93.27 | -0.195 | |
| BrrCDPK42 | Cluster-30924.119657 Cluster-30924.139625 | XM_013880416.3 | 1 791 | 596 | 66.84 | 8.98 | 81.88 | -0.355 | |
| BrrCDPK43 | Cluster-30924.119656 Cluster-30924.139627 | XM_013880416.3 | 1 791 | 596 | 66.91 | 8.98 | 81.39 | -0.360 | |
| BrrCDPK44 | Cluster-30924.139626 Cluster-30924.16433 | XM_013880416.3 | 1 791 | 596 | 66.87 | 8.98 | 81.22 | -0.356 | |
| BrrCDPK45 | Cluster-30924.139628 | XM_013880416.3 | 1 791 | 596 | 66.87 | 8.91 | 81.71 | -0.360 | |
| BrrCDPK46 | Cluster-30924.74759 | XM_013880416.3 | 1 791 | 596 | 66.85 | 8.98 | 81.88 | -0.354 | |
| BrrCDPK47 | Cluster-30924.114809 Cluster-30924.114810 Cluster-30924.123336 Cluster-30924.127303 Cluster-30924.130998 Cluster-30924.49496 | XM_048739418.1 | 1 743 | 580 | 65.01 | 8.91 | 80.88 | -0.359 | |
| BrrCDPK48 | Cluster-30924.101435 Cluster-30924.124946 | XM_048739418.1 | 1 743 | 580 | 65.00 | 8.91 | 80.71 | -0.363 | |
| BrrCDPK49 | Cluster-30924.155459 | XM_048769751.1 | 1 242 | 413 | 47.04 | 6.44 | 93.51 | -0.188 | |
| BrrCDPK50 | Cluster-30924.148411 | XM_048741021.1 | 1 785 | 594 | 66.10 | 8.76 | 82.64 | -0.314 | |
表3 芜菁CDPK基因家族的蛋白质理化性质分析
Table 3 Analysis of protein physicochemical properties of CDPK gene family in turnip
| 基因名 Gene name | 基因编号 Gene ID | 登录号 Accession number | cDNA长度/bp cDNA length | 氨基酸数 Number of amino acids | 分子量/kD Molecular weight | 等电点 Theoretical pI | 脂肪系数 Aliphatic index | 疏水性 GRAVY | |
|---|---|---|---|---|---|---|---|---|---|
| BrrCDPK1 | Cluster-30924.46777 | XM_009103718.3 | 1 737 | 578 | 64.58 | 8.95 | 83.39 | -0.317 | |
| BrrCDPK2 | Cluster-30924.137521 | XM_013807283.3 | 1 743 | 580 | 64.88 | 9.07 | 83.28 | -0.324 | |
| BrrCDPK3 | Cluster-30924.143914 | XM_013807283.3 | 1 731 | 576 | 64.47 | 9.01 | 83.68 | -0.318 | |
| BrrCDPK4 | Cluster-30924.13630 | XM_048738123.1 | 1 755 | 584 | 65.72 | 8.88 | 81.70 | -0.392 | |
| BrrCDPK5 | Cluster-30924.67984 | XM_010509132.2 | 1 806 | 601 | 67.46 | 8.99 | 88.80 | -0.318 | |
| BrrCDPK6 | Cluster-30924.158029 Cluster-30924.77034 Cluster-30924.77038 Cluster-30924.80376 | XM_009113894.3 | 1 785 | 594 | 66.74 | 9.06 | 83.43 | -0.374 | |
| BrrCDPK7 | Cluster-30924.58681 | XM_009113894.3 | 1 785 | 594 | 66.76 | 9.12 | 82.61 | -0.375 | |
| BrrCDPK8 | Cluster-30924.142390 | XM_009113894.3 | 1 785 | 594 | 66.77 | 9.06 | 83.60 | -0.373 | |
| BrrCDPK9 | Cluster-30924.157993 | XM_009113894.3 | 1 785 | 594 | 66.70 | 9.06 | 84.09 | -0.372 | |
| BrrCDPK10 | Cluster-30924.131630 | XM_013879035.3 | 1 704 | 567 | 63.43 | 8.84 | 86.84 | -0.218 | |
| BrrCDPK11 | Cluster-30924.136201 | XM_013792411.3 | 1 707 | 568 | 63.43 | 8.84 | 86.88 | -0.205 | |
| BrrCDPK12 | Cluster-30924.24510 | XM_013879035.3 | 1 704 | 567 | 63.59 | 8.67 | 85.47 | -0.219 | |
| BrrCDPK13 | Cluster-30924.57260 | XM_009140827.3 | 1 728 | 575 | 64.34 | 8.61 | 86.16 | -0.238 | |
| BrrCDPK14 | Cluster-30924.150034 | XM_013886734.3 | 1 713 | 570 | 63.80 | 8.85 | 86.74 | -0.251 | |
| BrrCDPK15 | Cluster-30924.150032 Cluster-30924.150033 | XM_009140827.3 | 1 728 | 575 | 64.38 | 8.70 | 86.16 | -0.243 | |
| BrrCDPK16 | Cluster-30924.14001 | XM_033289955.1 | 1 239 | 412 | 46.61 | 8.43 | 86.09 | -0.229 | |
| BrrCDPK17 | Cluster-30924.24231 | XM_013871943.3 | 1 236 | 411 | 46.25 | 8.00 | 88.42 | -0.203 | |
| BrrCDPK18 | Cluster-30924.82876 | XM_013895504.3 | 1 791 | 596 | 66.44 | 8.95 | 83.98 | -0.351 | |
| BrrCDPK19 | Cluster-30924.151434 | XM_013895504.3 | 1 791 | 596 | 66.45 | 8.88 | 84.80 | -0.343 | |
| BrrCDPK20 | Cluster-30924.142865 | XM_013895504.3 | 1 791 | 596 | 66.49 | 9.01 | 83.98 | -0.344 | |
| BrrCDPK21 | Cluster-30924.142866 | XM_009144190.3 | 1 791 | 596 | 66.45 | 8.95 | 83.98 | -0.356 | |
| BrrCDPK22 | Cluster-30924.146395 Cluster-30924.169091 | XM_013895504.3 | 1 791 | 596 | 66.44 | 8.88 | 83.98 | -0.350 | |
| BrrCDPK23 | Cluster-30924.52514 | XM_018622041.2 | 1 809 | 602 | 67.28 | 9.16 | 82.66 | -0.366 | |
| BrrCDPK24 | Cluster-30924.140928 Cluster-30924.155408 | XM_009144190.3 | 1 791 | 596 | 66.46 | 8.95 | 83.98 | -0.356 | |
| BrrCDPK25 | Cluster-30924.122791 | XM_013781701.1 | 1 734 | 577 | 64.40 | 8.52 | 84.85 | -0.260 | |
| BrrCDPK26 | Cluster-30924.24346 | XM_013892390.3 | 1 734 | 577 | 64.41 | 8.52 | 84.33 | -0.263 | |
| BrrCDPK27 | Cluster-30924.67985 | XM_009147578.3 | 1 983 | 660 | 74.20 | 9.02 | 85.41 | -0.248 | |
| BrrCDPK28 | Cluster-30924.67983 Cluster-30924.92828 | XM_009147578.3 | 1 983 | 660 | 74.31 | 9.07 | 86.15 | -0.246 | |
| BrrCDPK29 | Cluster-30924.11427 | XM_009147578.3 | 1 983 | 660 | 74.30 | 9.07 | 85.56 | -0.252 | |
| BrrCDPK30 | Cluster-30924.24345 | XM_009147578.3 | 1 983 | 660 | 74.33 | 9.07 | 84.97 | -0.254 | |
| BrrCDPK31 | Cluster-30924.77035 | XM_013732238.1 | 1 791 | 596 | 66.91 | 9.09 | 83.79 | -0.362 | |
| BrrCDPK32 | Cluster-30924.36432 | XM_013851068.3 | 1 791 | 596 | 66.95 | 9.13 | 83.79 | -0.355 | |
| BrrCDPK33 | Cluster-30924.77036 | XM_013893888.3 | 1 791 | 596 | 66.82 | 9.19 | 83.31 | -0.363 | |
| BrrCDPK34 | Cluster-30924.77037 | XM_013851068.3 | 1 791 | 596 | 66.91 | 9.13 | 83.79 | -0.362 | |
| BrrCDPK35 | Cluster-30924.114427 | XM_009147578.3 | 1 983 | 660 | 74.32 | 9.07 | 86.15 | -0.244 | |
| BrrCDPK36 | Cluster-69076.1 Cluster-69076.4 | XM_013786949.3 | 1 776 | 591 | 65.92 | 8.66 | 83.55 | -0.309 | |
| BrrCDPK37 | Cluster-69076.2 | XM_013786949.3 | 1 776 | 591 | 65.93 | 8.52 | 84.53 | -0.295 | |
| BrrCDPK38 | Cluster-69076.0 | XM_009151493.3 | 1 776 | 591 | 65.90 | 8.66 | 84.38 | -0.306 | |
| BrrCDPK39 | Cluster-30924.23018 | XM_013786949.3 | 1 776 | 591 | 65.95 | 8.77 | 83.89 | -0.314 | |
| BrrCDPK40 | Cluster-30924.12640 | XM_013787387.3 | 1 803 | 600 | 66.93 | 8.51 | 82.48 | -0.343 | |
| BrrCDPK41 | Cluster-30924.75342 | XM_048769751.1 | 1 242 | 413 | 47.04 | 6.68 | 93.27 | -0.195 | |
| BrrCDPK42 | Cluster-30924.119657 Cluster-30924.139625 | XM_013880416.3 | 1 791 | 596 | 66.84 | 8.98 | 81.88 | -0.355 | |
| BrrCDPK43 | Cluster-30924.119656 Cluster-30924.139627 | XM_013880416.3 | 1 791 | 596 | 66.91 | 8.98 | 81.39 | -0.360 | |
| BrrCDPK44 | Cluster-30924.139626 Cluster-30924.16433 | XM_013880416.3 | 1 791 | 596 | 66.87 | 8.98 | 81.22 | -0.356 | |
| BrrCDPK45 | Cluster-30924.139628 | XM_013880416.3 | 1 791 | 596 | 66.87 | 8.91 | 81.71 | -0.360 | |
| BrrCDPK46 | Cluster-30924.74759 | XM_013880416.3 | 1 791 | 596 | 66.85 | 8.98 | 81.88 | -0.354 | |
| BrrCDPK47 | Cluster-30924.114809 Cluster-30924.114810 Cluster-30924.123336 Cluster-30924.127303 Cluster-30924.130998 Cluster-30924.49496 | XM_048739418.1 | 1 743 | 580 | 65.01 | 8.91 | 80.88 | -0.359 | |
| BrrCDPK48 | Cluster-30924.101435 Cluster-30924.124946 | XM_048739418.1 | 1 743 | 580 | 65.00 | 8.91 | 80.71 | -0.363 | |
| BrrCDPK49 | Cluster-30924.155459 | XM_048769751.1 | 1 242 | 413 | 47.04 | 6.44 | 93.51 | -0.188 | |
| BrrCDPK50 | Cluster-30924.148411 | XM_048741021.1 | 1 785 | 594 | 66.10 | 8.76 | 82.64 | -0.314 | |
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
|
陈湘宏, 刘燕, 翁裕馨, 康文娟, 杨仕兵. 2014. 芜菁挥发油对高脂高糖小鼠降血糖的作用机制. 山东大学学报(医学版),(12):20-23.
|
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
|
费小钰, 李红丽, 王俊皓. 2017. 植物钙依赖蛋白激酶CDPK基因功能综述. 吉林农业,(9):104-105.
|
|
| [10] |
|
| [11] |
|
|
龚丽莎, 向芷萱, 王照, 鲁敏, 安华明. 2023. 刺梨CDPK基因家族的鉴定及其对供钙水平的表达响应. 果树学报, 40 (4):639-652.
|
|
| [12] |
|
|
靳燕, 王谢琴, 卿华, 马政, 姚慧鹏. 2021. 黄瓜CDPK基因家族的鉴定与进化特征分析. 四川农业大学学报, 39 (1):19-26.
|
|
| [13] |
doi: 10.1073/pnas.1214808109 pmid: 23112190 |
| [14] |
doi: 10.1186/1471-2164-14-433 pmid: 23815483 |
| [15] |
doi: 10.7505/j.issn.1007-9084.2016.03.004 |
|
李可琪, 王莎莎, 曾新华, 闫晓红, 吴刚. 2016. 甘蓝型油菜温敏GMSTE5A幼蕾酵母双杂交cDNA文库的构建及筛选. 中国油料作物学报, 38 (3):292-299.
doi: 10.7505/j.issn.1007-9084.2016.03.004 |
|
| [16] |
doi: 10.1046/j.1365-313x.2001.01008.x pmid: 11359610 |
| [17] |
doi: S0024-3205(18)30591-5 pmid: 30248349 |
| [18] |
doi: S1569-9048(18)30243-X pmid: 30447305 |
| [19] |
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0719 |
|
李亚男, 张豪杰, 梁梦静, 罗涛, 李旺宁, 张春辉, 季春丽, 李润植, 薛金爱, 崔红利. 2024. 雨生红球藻钙依赖蛋白激酶(CDPK)家族鉴定与表达分析. 生物技术通报, 40 (2):300-312.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0719 |
|
| [20] |
doi: 10.11686/cyxb2022386 |
|
刘玉玲, 朱新霞, 吕新华, 孙辉. 2023. 雪莲SikCDPK1基因的表达特征和蛋白激酶活性分析. 草业学报, 32 (9):213-221.
doi: 10.11686/cyxb2022386 |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
doi: 10.1093/emboj/20.20.5556 pmid: 11597999 |
| [25] |
doi: 10.1104/pp.000869 pmid: 12011347 |
| [26] |
|
| [27] |
|
| [28] |
doi: 10.1105/tpc.109.066936 pmid: 19880793 |
| [29] |
|
| [30] |
|
|
王文宁. 2018. 蔓菁营养成分分析及其改善小鼠肠道菌群的研究[硕士论文]. 郑州: 郑州大学.
|
|
| [31] |
pmid: 17873090 |
| [32] |
|
|
张海鑫. 2022. 多毛番茄CDPK基因家族分析及胁迫响应分析[硕士论文]. 哈尔滨: 东北农业大学.
|
|
| [33] |
|
| [34] |
|
| [35] |
|
|
张习敏. 2018. 高钙诱导天蓝苜蓿叶片草酸钙积累及其在环境适应中的作用研究[博士论文]. 厦门: 厦门大学.
|
|
| [36] |
|
| [37] |
|
|
庄丽丽, 宋林眉, 王芳, 李玉群, 张柱岐, 刘凯. 2023. 核桃CDPK基因家族鉴定与转录表达分析. 落叶果树, 55 (6):32-35.
|
| [1] | 马曦, 张金睿, 庄红梅, 陈巍, 张蜜, 袁晓琪, 易小芳, 王聪聪, 王海云, 王燕. 发根农杆菌介导的芜菁高效遗传转化体系建立[J]. 园艺学报, 2025, 52(5): 1389-1398. |
| [2] | 付琪, 王丹, 景维坤, 张颢, 王慧纯, 蹇洪英, 邱显钦, 王其刚, 唐开学, 晏慧君. 月季类胡萝卜素裂解双加氧酶基因RcCCD4在花香合成中的功能[J]. 园艺学报, 2025, 52(3): 623-634. |
| [3] | 谢丽华, 姚鹏强, 刘佳雪, 王哲, 朱楠楠, 程世平, 程占超. 毛竹PheNAC4互作蛋白的筛选与分析[J]. 园艺学报, 2025, 52(10): 2677-2690. |
| [4] | 梁国平, 曾宝珍, 刘铭, 边志远, 陈佰鸿, 毛娟. 山葡萄VaSR基因家族的鉴定及VaSR1抗寒功能验证与互作蛋白筛选[J]. 园艺学报, 2025, 52(1): 37-50. |
| [5] | 赵佳莹, 曾周婷, 岑欣颖, 施姣淇, 李效贤, 沈晓霞, 俞振明. 铁皮石斛CCO基因家族鉴定及其在花发育中的表达分析[J]. 园艺学报, 2024, 51(9): 2075-2088. |
| [6] | 邱辉, 朱德娟, 张永乐, 高玉洁, 李柳, 王国平, 洪霓. ACLSV外壳蛋白与梨两种E3泛素连接酶互作及亚细胞定位[J]. 园艺学报, 2024, 51(8): 1715-1727. |
| [7] | 段敏杰, 李怡斐, 王春萍, 杨小苗, 黄任中, 黄启中, 张世才. 辣椒果实类胡萝卜素调控因子转录组和靶向代谢组分析[J]. 园艺学报, 2024, 51(8): 1773-1791. |
| [8] | 杨亮, 刘欢, 马燕勤, 李菊, 王海娥, 周玉洁, 龙海成, 苗明军, 李志, 常伟. 利用CRISPR/Cas9技术创制高番茄红素番茄新材料[J]. 园艺学报, 2024, 51(2): 253-265. |
| [9] | 蒋政华, 刘胜军, 刘文洁, 魏冉冉, 何政辰, 程超, 唐政, 刘保艳, 谢宗周, 叶俊丽, 柴利军, 岳鹏涛, 邓秀新. 基于Pull-Down/MS技术筛选柑橘CsMYC2和CsMPK6的互作蛋白[J]. 园艺学报, 2024, 51(12): 2913-2927. |
| [10] | 汪书杰, 栾雨婷, 徐昌杰. 植物叶黄素酯化研究进展[J]. 园艺学报, 2023, 50(9): 1830-1840. |
| [11] | 田密霞, 周福慧, 姜爱丽, 祝朋芳, 陈晨, 刘程惠, 原畅. 芸薹属植物呈色机理研究进展[J]. 园艺学报, 2023, 50(9): 1971-1986. |
| [12] | 肖翔, 周储江, 金舒婉, 施丽愉, 杨震峰, 曹士锋, 陈伟. PpMADS2与PpMADS3协同调控黄肉桃果实类胡萝卜素积累机制的研究[J]. 园艺学报, 2023, 50(6): 1173-1186. |
| [13] | 杜艳霞, 王艺光, 肖政, 董彬, 方遒, 钟诗蔚, 杨丽媛, 赵宏波. 桂花OfNCED3调控转基因烟草叶片类胡萝卜素和叶绿素的合成[J]. 园艺学报, 2023, 50(6): 1284-1294. |
| [14] | 俞沁含, 李俊铎, 崔莹, 王佳慧, 郑巧玲, 徐伟荣. 山葡萄转录因子VaMYB4a互作蛋白的筛选与鉴定[J]. 园艺学报, 2023, 50(3): 508-522. |
| [15] | 丁磊, 孙平平, 张磊, 李正男. 利用Y2H筛选西方烟中与苹果茎痘病毒CP互作的蛋白[J]. 园艺学报, 2023, 50(11): 2509-2515. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司