
园艺学报 ›› 2023, Vol. 50 ›› Issue (6): 1215-1229.doi: 10.16420/j.issn.0513-353x.2022-0339
杨孟霞, 刘晓林, 曹雪, 魏凯, 宁宇, 杨沛, 李珊珊, 陈紫月, 王孝宣, 国艳梅, 杜永臣, 李君明, 刘磊, 李鑫, 黄泽军( )
)
收稿日期:2022-11-29
修回日期:2023-02-27
出版日期:2023-06-25
发布日期:2023-06-27
通讯作者:
* (E-mail:huangzejun@caas.cn)基金资助:
YANG Mengxia, LIU Xiaolin, CAO Xue, WEI Kai, NING Yu, YANG Pei, LI Shanshan, CHEN Ziyue, WANG Xiaoxuan, GUO Yanmei, DU Yongchen, LI Junming, LIU Lei, LI Xin, HUANG Zejun( )
)
Received:2022-11-29
Revised:2023-02-27
Published:2023-06-25
Online:2023-06-27
摘要:
为了构建番茄CRISPR/Cas9介导的多基因编辑体系,用SlU6-2p、SlU6-3p、SlU6-7p、SlU3-5p、SlU3-9p和SlU6-5p启动子分别替换pKSE401和pCBC-DT1T2载体中的拟南芥U6启动子,构建了1个双元载体(pMGET)、2个中间载体(pKC-S2M和pKC-S3M)和3个gRNA模块载体(pCBC-S1、pCBC-S2和pCBC-S3)。为了测试该多基因编辑体系,通过PCR扩增、Golden gate克隆和同尾酶技术,将含有6个番茄果实性状相关基因Green flesh(GF)、Ovate(O)、Locule number(LC)、SlMYB12(Y)、Tangerine(T)、Uniform ripening(U)靶点序列的sgRNA聚合构建多基因编辑载体pMGET-OYGTULC和pMGET-TULCOYG,检测6个基因同时被编辑的效率分别为44.00%和11.76%。用携带pMGET-OYGTULC载体的根癌农杆菌转化番茄材料获得2株6个基因均被编辑的植株,序列变异为单个碱基的插入、单个或多个碱基的缺失及大片段的缺失等多种形式。本研究中该多基因编辑体系可以高效地应用于番茄多基因编辑,并且可以得到稳定遗传的突变体,可为番茄基础研究和遗传改良提供一个简便的工具箱。
中图分类号:
杨孟霞, 刘晓林, 曹雪, 魏凯, 宁宇, 杨沛, 李珊珊, 陈紫月, 王孝宣, 国艳梅, 杜永臣, 李君明, 刘磊, 李鑫, 黄泽军. 番茄CRISPR/Cas9介导的多基因编辑技术体系构建与应用[J]. 园艺学报, 2023, 50(6): 1215-1229.
YANG Mengxia, LIU Xiaolin, CAO Xue, WEI Kai, NING Yu, YANG Pei, LI Shanshan, CHEN Ziyue, WANG Xiaoxuan, GUO Yanmei, DU Yongchen, LI Junming, LIU Lei, LI Xin, HUANG Zejun. Construction and Application of a CRISPR/Cas9 System for Multiplex Gene Editing in Tomato[J]. Acta Horticulturae Sinica, 2023, 50(6): 1215-1229.
| 引物Primer | 引物序列(5′-3′)Primer sequence |
|---|---|
| MGET-17 | TTCATCTCGGGTCTCGTTTGATCCACGATGATCTCCGTTGTTTTAGAGCTAGAAATAG |
| MGET-18 | TTCATCTCGGGTCTCtAAACTCTGCCTCTCTTGATGCCCTGACCGTTGATAGTGGATAG |
| MGET-19 | TTCATCTCGGGTCTCGCTTGTCCATTGCCACATTAGTGGGTTTTAGAGCTAGAAATAG |
| MGET-20 | TTCATCTCGGGTCTCtAAACCCCAACTCAAGCCACAAAGTGACATATCCTACTGGTACG |
| MGET-21 | TTCATCTCGGGTCTCGTTTGGTGAAGAACCTGCTATTCAGTTTTAGAGCTAGAAATAG |
| MGET-43 | TTCATCTCGGGTCTCtAAACATTAGTAAACATCATGCCACAAACACACATGCTAATATTC |
| MGET-23 | TTCATCTCGGGTCTCGTTTGCTTTGTGGCTTGAGTTGGGGTTTTAGAGCTAGAAATAG |
| MGET-24 | TTCATCTCGGGTCTCtAAACTGAATAGCAGGTTCTTCACTGACCGTTGATAGTGGATAG |
| MGET-44 | TTCATCTCGGGTCTCGCTTGTGGCATGATGTTTACTAATGTTTTAGAGCTAGAAATAG |
| MGET-26 | TTCATCTCGGGTCTCtAAACAACGGAGATCATCGTGGATTGACATATCCTACTGGTACG |
| MGET-27 | TTCATCTCGGGTCTCGTTTGGGGCATCAAGAGAGGCAGAGTTTTAGAGCTAGAAATAG |
| MGET-28 | TTCATCTCGGGTCTCtAAACCCACTAATGTGGCAATGGACAAACACACATGCTAATATTC |
| ZP121 | GAACAAAAGCCTTGAGAACC |
| ZP122 | GTCGAAATCCCACACATCAC |
| ZP123 | CAACTAACCTTATCACTGAATCC |
| ZP124 | TGGTTAACTTTCCAGGGAGC |
| ZP125 | GATGCTGGCATTTGTGTGATTC |
| ZP126 | ACGACTAGAACCGTCAACGA |
| ZP129 | CAAAGAGTGACATGCAATGC |
| ZP130 | CCTCTTGAGAAGTAATGTTCC |
| ZP149 | GCAGAGACAAAAAGAATGGG |
| ZP150 | CATGATTAATCAAAGGAGGTTC |
| ZP191 | GCCGAACACATCAACATTTC |
| ZP192 | CCACACGTCTCGTCAGAATG |
| Cas9-909F | GCAGCTCTCCAAGGACACAT |
| Cas9-2450R | CGTGAGTTCTTCTGGCCCTT |
表 1 主要引物序列信息
Table 1 Sequence information of primers
| 引物Primer | 引物序列(5′-3′)Primer sequence |
|---|---|
| MGET-17 | TTCATCTCGGGTCTCGTTTGATCCACGATGATCTCCGTTGTTTTAGAGCTAGAAATAG |
| MGET-18 | TTCATCTCGGGTCTCtAAACTCTGCCTCTCTTGATGCCCTGACCGTTGATAGTGGATAG |
| MGET-19 | TTCATCTCGGGTCTCGCTTGTCCATTGCCACATTAGTGGGTTTTAGAGCTAGAAATAG |
| MGET-20 | TTCATCTCGGGTCTCtAAACCCCAACTCAAGCCACAAAGTGACATATCCTACTGGTACG |
| MGET-21 | TTCATCTCGGGTCTCGTTTGGTGAAGAACCTGCTATTCAGTTTTAGAGCTAGAAATAG |
| MGET-43 | TTCATCTCGGGTCTCtAAACATTAGTAAACATCATGCCACAAACACACATGCTAATATTC |
| MGET-23 | TTCATCTCGGGTCTCGTTTGCTTTGTGGCTTGAGTTGGGGTTTTAGAGCTAGAAATAG |
| MGET-24 | TTCATCTCGGGTCTCtAAACTGAATAGCAGGTTCTTCACTGACCGTTGATAGTGGATAG |
| MGET-44 | TTCATCTCGGGTCTCGCTTGTGGCATGATGTTTACTAATGTTTTAGAGCTAGAAATAG |
| MGET-26 | TTCATCTCGGGTCTCtAAACAACGGAGATCATCGTGGATTGACATATCCTACTGGTACG |
| MGET-27 | TTCATCTCGGGTCTCGTTTGGGGCATCAAGAGAGGCAGAGTTTTAGAGCTAGAAATAG |
| MGET-28 | TTCATCTCGGGTCTCtAAACCCACTAATGTGGCAATGGACAAACACACATGCTAATATTC |
| ZP121 | GAACAAAAGCCTTGAGAACC |
| ZP122 | GTCGAAATCCCACACATCAC |
| ZP123 | CAACTAACCTTATCACTGAATCC |
| ZP124 | TGGTTAACTTTCCAGGGAGC |
| ZP125 | GATGCTGGCATTTGTGTGATTC |
| ZP126 | ACGACTAGAACCGTCAACGA |
| ZP129 | CAAAGAGTGACATGCAATGC |
| ZP130 | CCTCTTGAGAAGTAATGTTCC |
| ZP149 | GCAGAGACAAAAAGAATGGG |
| ZP150 | CATGATTAATCAAAGGAGGTTC |
| ZP191 | GCCGAACACATCAACATTTC |
| ZP192 | CCACACGTCTCGTCAGAATG |
| Cas9-909F | GCAGCTCTCCAAGGACACAT |
| Cas9-2450R | CGTGAGTTCTTCTGGCCCTT |
| 基因名称 Gene name | 基因ID Gene ID | 靶点序列(5′-3′) Target sequence | PAM区 PAM area |
|---|---|---|---|
| Green flesh(GF) | Solyc08g080090 | CCACTAATGTGGCAATGGA | CCT |
| Ovate(O) | Solyc02g085500 | ATCCACGATGATCTCCGTT | GGG |
| Locule number(LC) | Solyc02g083950 | ATTAGTAAACATCATGCCA | CCA |
| Tangerine(T) | Solyc10g081650 | CCCAACTCAAGCCACAAAG | CCC |
| Uniform(U) | Solyc10g008160 | GTGAAGAACCTGCTATTCA | TGG |
| SlMYB12(Y) | Solyc01g079620 | GGGCATCAAGAGAGGCAGA | TGG |
表2 多基因敲除的靶点核苷酸序列
Table 2 The target nucleotide sequence of multiple genes
| 基因名称 Gene name | 基因ID Gene ID | 靶点序列(5′-3′) Target sequence | PAM区 PAM area |
|---|---|---|---|
| Green flesh(GF) | Solyc08g080090 | CCACTAATGTGGCAATGGA | CCT |
| Ovate(O) | Solyc02g085500 | ATCCACGATGATCTCCGTT | GGG |
| Locule number(LC) | Solyc02g083950 | ATTAGTAAACATCATGCCA | CCA |
| Tangerine(T) | Solyc10g081650 | CCCAACTCAAGCCACAAAG | CCC |
| Uniform(U) | Solyc10g008160 | GTGAAGAACCTGCTATTCA | TGG |
| SlMYB12(Y) | Solyc01g079620 | GGGCATCAAGAGAGGCAGA | TGG |
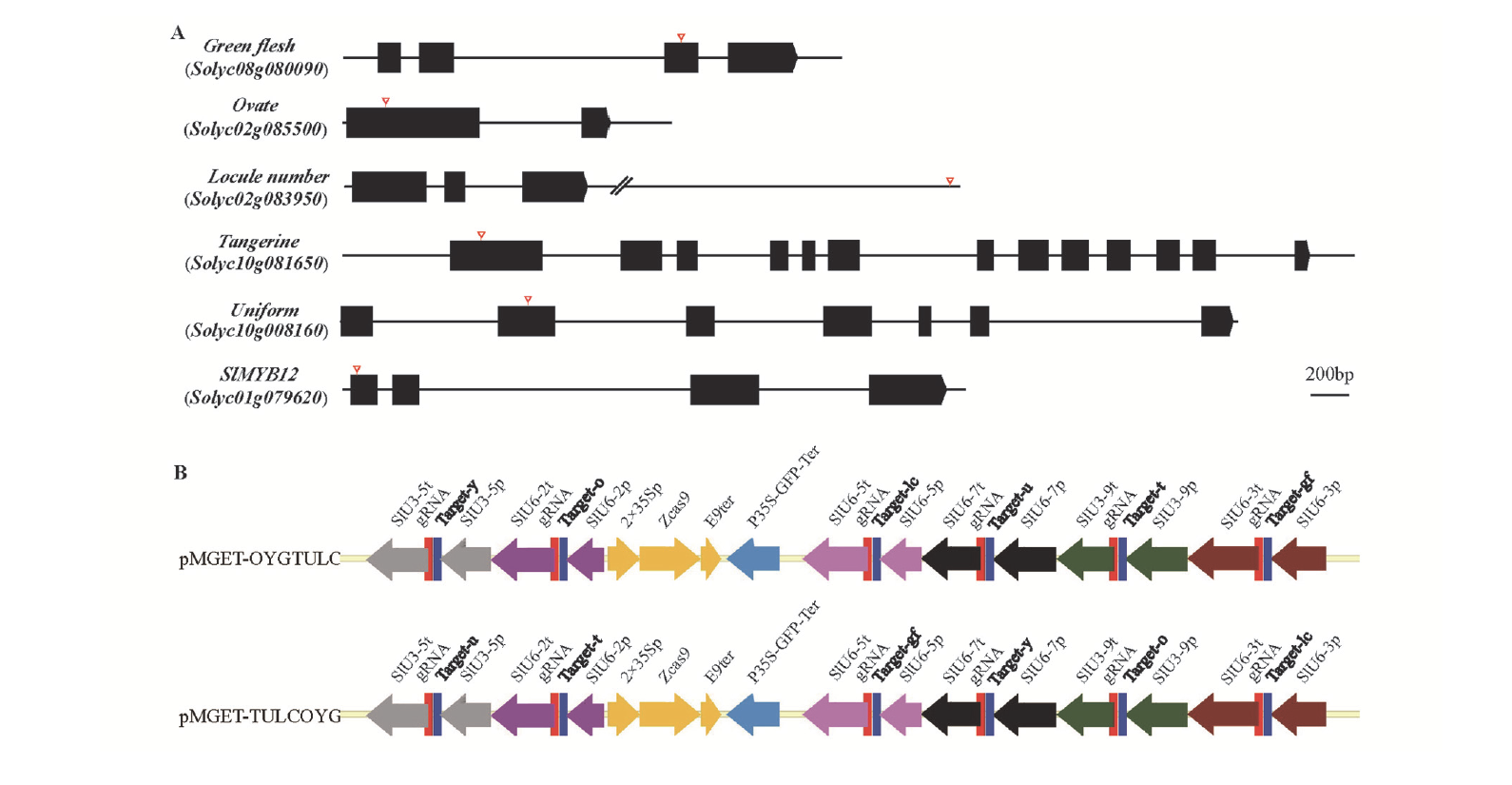
图3 基因编辑载体结构和基因靶点位置示意图 A:GF、O、LC、T、U、Y基因及靶点位置示意图,红色三角表示靶点位置;B:不同启动子驱动sgRNA的载体示意图。
Fig. 3 Diagram of gene target location and gene editing vector structure A:Represents intron-exon structure of GF,O,LC,T,U and Y gene and the location of their target sites,and the red triangle indicates the target site;B:Represents the vector diagram of sgRNA driven by different promoters.
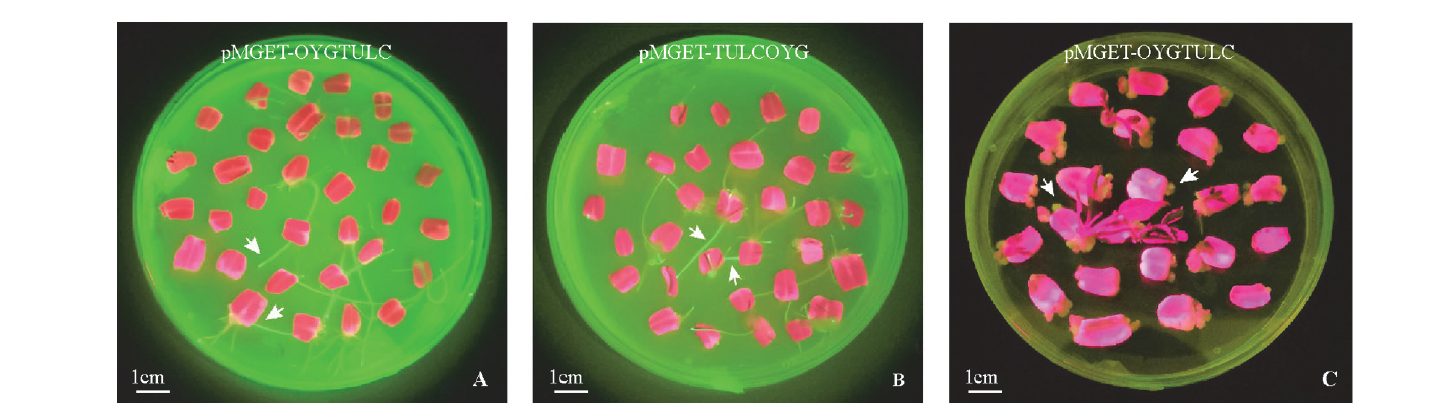
图4 含基因编辑载体农杆菌诱导的番茄子叶毛状根(A、B)和愈伤组织(C)的绿色荧光图
Fig. 4 Green fluorescence spectra of hairy roots(A,B)and callus(C)of tomato cotyledons induced by Agrobacterium containing gene editing vector
| 载体名称 Vector name | 基因名称 Gene name | 启动子 Promoter | 测序的毛状根数 Number of roots for sequencing | 单基因编辑 Single gene editing | 6个基因同时编辑 Six genes are edited at the same time | ||
|---|---|---|---|---|---|---|---|
| 数量Number | 效率/% Efficiency | 数量Number | 效率/% Efficiency | ||||
| pMGET- | Green flesh | SlU6-3p | 50 | 45 | 83.33 | ||
| OYGTULC | Tangerine | SlU3-9p | 50 | 41 | 75.93 | ||
| Uniform | SlU6-7p | 50 | 39 | 72.22 | |||
| SlMYB12 | SlU3-5p | 50 | 45 | 83.33 | |||
| Ovate | SlU6-2p | 50 | 29 | 53.70 | |||
| Locule number | SlU6-5p | 50 | 45 | 83.33 | |||
| 小计 Subtotal | 22 | 44.00 | |||||
| pMGET- | Green flesh | SlU6-5p | 51 | 49 | 96.08 | ||
| TULCOYG | Tangerine | SlU6-2p | 51 | 50 | 98.04 | ||
| Uniform | SlU3-5p | 51 | 10 | 19.61 | |||
| SlMYB12 | SlU6-7p | 51 | 49 | 96.08 | |||
| Ovate | SlU3-9p | 51 | 38 | 74.51 | |||
| Locule number | SlU6-3p | 51 | 50 | 98.04 | |||
| 小计 Subtotal | 6 | 11.76 | |||||
表3 两个载体不同基因的编辑效率
Table 3 Editing efficiency of different genes of two vectors
| 载体名称 Vector name | 基因名称 Gene name | 启动子 Promoter | 测序的毛状根数 Number of roots for sequencing | 单基因编辑 Single gene editing | 6个基因同时编辑 Six genes are edited at the same time | ||
|---|---|---|---|---|---|---|---|
| 数量Number | 效率/% Efficiency | 数量Number | 效率/% Efficiency | ||||
| pMGET- | Green flesh | SlU6-3p | 50 | 45 | 83.33 | ||
| OYGTULC | Tangerine | SlU3-9p | 50 | 41 | 75.93 | ||
| Uniform | SlU6-7p | 50 | 39 | 72.22 | |||
| SlMYB12 | SlU3-5p | 50 | 45 | 83.33 | |||
| Ovate | SlU6-2p | 50 | 29 | 53.70 | |||
| Locule number | SlU6-5p | 50 | 45 | 83.33 | |||
| 小计 Subtotal | 22 | 44.00 | |||||
| pMGET- | Green flesh | SlU6-5p | 51 | 49 | 96.08 | ||
| TULCOYG | Tangerine | SlU6-2p | 51 | 50 | 98.04 | ||
| Uniform | SlU3-5p | 51 | 10 | 19.61 | |||
| SlMYB12 | SlU6-7p | 51 | 49 | 96.08 | |||
| Ovate | SlU3-9p | 51 | 38 | 74.51 | |||
| Locule number | SlU6-3p | 51 | 50 | 98.04 | |||
| 小计 Subtotal | 6 | 11.76 | |||||
| [1] |
Barry C S, McQuinn R P, Chung M Y, Besuden A, Giovannoni J J. 2008. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol, 147 (1):179-187.
doi: 10.1104/pp.108.118430 pmid: 18359841 |
| [2] |
Brooks C, Nekrasov V, Lippman Z B, van Eck J. 2014. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 system. Plant Physiol, 166 (3):1292-1297.
doi: 10.1104/pp.114.247577 URL |
| [3] |
Čermák T, Curtin S J, Gil-Humanes J, Čegan R, Kono T J Y, Konečná E, Belanto J J, Starker C G, Mathre J W, Greenstein R L, Voytas D F. 2017. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29 (6):1196-1217.
doi: 10.1105/tpc.16.00922 URL |
| [4] | Chaudhary J, Alisha A, Bhatt V, Chandanshive S, Kumar N, Mir Z, Kumar A, Yadav S K, Shivaraj S M, Sonah H, Deshmukh R. 2019. Mutation breeding in tomato:Advances,applicability and challenges. Plants(Basel), 8 (5):128. |
| [5] |
Chen K, Gao C. 2014. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Reports, 33 (4):575-583.
doi: 10.1007/s00299-013-1539-6 pmid: 24277082 |
| [6] | Cui Xia, Zhang Shuai-bin. 2017. The utilization and prospect of genome editing in horticultural crops. Acta Horticulturae Sinica, 44 (9):1787-1795. (in Chinese) |
| 崔霞, 张率斌. 2017. 基因编辑技术及其在园艺作物中的应用和展望. 园艺学报, 44 (9):1787-1795. | |
| [7] |
Deng L, Wang H, Sun C, Li Q, Jiang H, Du M, Li C B, Li C. 2018. Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J Genet Genomics, 45 (1):51-54.
doi: S1673-8527(17)30178-9 pmid: 29157799 |
| [8] | Du H Y, Zeng X R, Zhao M, Cui X P, Wang Q, Yang H, Cheng H, Yu D Y. 2016. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. Biotechnol, 217:90-97. |
| [9] |
Du M, Zhou K, Liu Y, Deng L, Zhang X, Lin L, Zhou M, Zhao W, Wen C, Xing J, Li C B, Li C. 2020. A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J, 102 (5):1090-1100.
doi: 10.1111/tpj.v102.5 URL |
| [10] |
Gao Y, Zhu N, Zhu X, Wu M, Jiang C Z, Grierson D, Luo Y, Shen W, Zhong S, Fu D Q, Qu G. 2019. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas 9 CNR and NOR mutants. Hortic Res, 6:39.
doi: 10.1038/s41438-019-0122-x |
| [11] |
Hisamatsu S, Sakaue M, Takizawa A, Kato T, Kamoshita M, Ito J, Kashiwazaki N. 2015. Knockout of targeted gene in porcine somatic cells using zinc-finger nuclease. Anim Sci J, 86 (2):132-137.
doi: 10.1111/asj.12259 pmid: 25187232 |
| [12] |
Isaacson T, Ronen G, Zamir D, Hirschberg J. 2002. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell, 14 (2):333-342.
doi: 10.1105/tpc.010303 pmid: 11884678 |
| [13] |
Kimura S, Sinha N. 2008. Tomato(Solanum lycopersicum):a model fruit-bearing crop. CSH Protocols, DOI:10.1101/pdb.emo105.
doi: 10.1101/pdb.emo105 |
| [14] |
Kiss T, Solymosy F. 1990. Molecular analysis of a U3 RNA gene locus in tomato:transcription signals,the coding region,expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res, 18 (8):1941-1949.
pmid: 2336383 |
| [15] |
Klap C, Yeshayahou E, Bolger A M, Arazi T, Gupta S K, Shabtai S, Usadel B, Salts Y, Barg R. 2017. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J, 15 (5):634-647.
doi: 10.1111/pbi.2017.15.issue-5 URL |
| [16] |
Kwon C T, Heo J, Lemmon Z H, Capua Y, Hutton S F, van Eck J, Park S J, Lippman Z B. 2020. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat Biotechnol, 38 (2):182-188.
doi: 10.1038/s41587-019-0361-2 |
| [17] |
Li R, Li R, Li X, Fu D, Zhu B, Tian H, Luo Y, Zhu H. 2018a. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol J, 16 (2):415-427.
doi: 10.1111/pbi.2018.16.issue-2 URL |
| [18] |
Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C. 2018b. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol, 36:1160-1163.
doi: 10.1038/nbt.4273 URL |
| [19] |
Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y, Zhu H. 2018c. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci, 9:559.
doi: 10.3389/fpls.2018.00559 URL |
| [20] |
Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, Huang Z, Li J, Zhang C, Wang T, Zhang Y, Wang A, Zhang Y, Lin K, Li C, Xiong G, Xue Y, Mazzucato A, Causse M, Fei Z, Giovannoni J J, Chetelat R T, Zamir D, Städler T, Li J, Ye Z, Du Y, Huang S. 2014. Genomic analyses provide insights into the history of tomato breeding. Nat Genet, 46 (11):1220-1226.
doi: 10.1038/ng.3117 pmid: 25305757 |
| [21] | Liu Chun-xia, Gui Hua-ping, Wen Qin, Liu Yu-bo, Gao Yang, Zhang Xiu-juan, Cui Rui-jie, Xu Jian-ping. 2020. CRISPR/Cas 9 genome editing system optimization in tomato. Molecular Plant Breeding, 18 (20):6716-6724. (in Chinese) |
| 刘春霞, 桂花萍, 文琴, 刘宇博, 高阳, 张秀娟, 崔瑞洁, 许建平. 2020. 番茄CRISPR/Cas9基因编辑效率的优化. 分子植物育种, 18 (20):6716-6724. | |
| [22] |
Liu J, van Eck J, Cong B, Tanksley S D. 2002. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci U S A, 99 (20):13302-13306.
doi: 10.1073/pnas.162485999 URL |
| [23] |
Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, Lu Y, Wang Q, Li C, Zhai Q. 2019. MYC 2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell, 31 (1):106-127.
doi: 10.1105/tpc.18.00405 URL |
| [24] |
Lu J, Pan C, Li X, Huang Z, Shu J, Wang X, Lu X, Pan F, Hu J, Zhang H, Su W, Zhang M, Du Y, Liu L, Guo Y, Li J. 2021. OBV(obscure vein),a C2H 2 zinc finger transcription factor,positively regulates chloroplast development and bundle sheath extension formation in tomato(Solanum lycopersicum)leaf veins. Hortic Res, 8 (1):230.
doi: 10.1038/s41438-021-00659-z |
| [25] |
Ma X L, Zhang Q Y, Zhu Q L, Liu W, Chen Y, Qiu R, Wang B, Yang Z F, Li H Y, Lin Y R, Xie Y Y, Shen R X, Chen S F, Wang Z, Chen Y L, Guo J X, Chen L T, Zhao X C, Dong Z C, Liu Y G. 2015. A robust CRISPR/Cas 9 system for convenient,high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant, 8 (8):1274-1284.
doi: 10.1016/j.molp.2015.04.007 URL |
| [26] |
Montecillo J A V, Chu L L, Bae H. 2020. CRISPR-Cas 9 system for plant genome editing:current approaches and emerging developments. Agronomy, 10 (7):1033.
doi: 10.3390/agronomy10071033 URL |
| [27] |
Muños S, Ranc N, Botton E, Bérard A, Rolland S, Duffé P, Carretero Y, Le Paslier M C, Delalande C, Bouzayen M, Brunel D, Causse M. 2011. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol, 156 (4):2244-2254.
doi: 10.1104/pp.111.173997 pmid: 21673133 |
| [28] |
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. 2017. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep, 7 (1):482.
doi: 10.1038/s41598-017-00578-x pmid: 28352080 |
| [29] |
Nonaka S, Arai C, Takayama M, Matsukura C, Ezura H. 2017. Efficient increase of ɣ-aminobutyric acid(GABA)content in tomato fruits by targeted mutagenesis. Sci Rep, 7 (1):7057.
doi: 10.1038/s41598-017-06400-y |
| [30] |
Powell A L, Nguyen C V, Hill T, Cheng K L, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, Lopez-Baltazar J, Barry C S, Liu Y, Chetelat R, Granell A, van Deynze A, Giovannoni J J, Bennett A B. 2012. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science, 336 (6089):1711-1715.
doi: 10.1126/science.1222218 pmid: 22745430 |
| [31] |
Rodríguez-Leal D, Lemmon Z H, Man J, Bartlett M E, Lippman Z B. 2017. Engineering quantitative trait variation for crop improvement by genome editing. Cell, 171 (2):470-480.
doi: S0092-8674(17)30988-1 pmid: 28919077 |
| [32] |
Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso M A, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, Federici F, Sinha N, Deal R B, Bailey-Serres J, Brady S M. 2014. Hairy root transformation using Agrobacterium rhizo genes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol, 166 (2):455-469.
doi: 10.1104/pp.114.239392 |
| [33] |
Shan Qi-wei, Gao Cai-xia. 2015. Research progress of genome editing and derivative technologies in plants. Hereditas(Beijing), 37 (10):953-973. (in Chinese)
doi: 10.16288/j.yczz.15-156 pmid: 26496748 |
| 单奇伟, 高彩霞. 2015. 植物基因组编辑及衍生技术最新研究进展. 遗传, 37 (10):953-973. | |
| [34] |
Sun N, Zhao H. 2013. Transcription activator-like effector nucleases(TALENs):A highly efficient and versatile tool for genome editing. Biotechnol Bioeng, 110 (7):1811-1821.
doi: 10.1002/bit.24890 URL |
| [35] |
Sun X J, Hu Z, Chen R, Jiang Q Y, Song G H, Zhang H, Xi Y J. 2015. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep, 5:10342.
doi: 10.1038/srep10342 pmid: 26022141 |
| [36] | van der Knaap E, Chakrabarti M, Chu Y H, Clevenger J P, Illa-Berenguer E, Huang Z, Keyhaninejad N, Mu Q, Sun L, Wang Y, Wu S. 2014. What lies beyond the eye:the molecular mechanisms regulating tomato fruit weight and shape. Front Plant Sci, 5:227. |
| [37] |
Waibel F, Filipowicz W. 1990. U6 snRNA genes of Arabidopsis are transcribed by RNA polymerase III but contain the same two upstream promoter elements as RNA polymerase II-transcribed U-snRNA genes. Nucleic Acids Res, 18 (12):3451-3458.
pmid: 2362802 |
| [38] |
Wang C, Shen L, Fu Y, Yan C, Wang K. 2015. A simple CRISPR/Cas 9 system for multiplex genome editing in rice. J Genet Genomics, 42 (12):703-706.
doi: 10.1016/j.jgg.2015.09.011 URL |
| [39] | Wang Dan, Wang Mi, Liu Jun, Zhou Xiaohui, Liu Songyu, Yang Yan, Zhuang Yong. 2022. Cloning of U6 promoters and establishment of CRISPR/Cas 9 mediated gene editing system in eggplant. Acta Horticulturae Sinica, 49 (4):791-800. (in Chinese) |
|
王丹, 王谧, 刘军, 周晓慧, 刘松瑜, 杨艳, 庄勇. 2022. 茄子U6启动子克隆及CRISPR/Cas9介导的基因编辑体系建立. 园艺学报, 49 (4):791-800.
doi: 10.16420/j.issn.0513-353x.2020-0265 URL |
|
| [40] | Wang Juan, Wang Baike, Li Ning, Yang Tao, Patiguli ASMUTOLA, Yu Qinghui. 2020. Application progress of gene editing technology in tomato breeding. Plant Physiology Journal, 56 (12):2606-2616. (in Chinese) |
| 王娟, 王柏柯, 李宁, 杨涛, 帕提古丽 · 艾斯木托拉, 余庆辉. 2020. 基因编辑技术在番茄育种中的应用进展. 植物生理学报, 56 (12):2606-2616. | |
| [41] | Wang Mu-jia, Zhu Jian-kang. 2018. Simplification of multiplex gene editing systems in plants. Chinese Bulletin of Life Sciences, 30 (9):980-986. (in Chinese) |
| 王木桂, 朱健康. 2018. 化繁为简——植物多基因编辑体系的优化. 生命科学, 30 (9):980-986. | |
| [42] |
Xing H L, Dong L, Wang Z P, Zhang H Y, Han C Y, Liu B, Wang X C, Chen Q J. 2014. A CRISPR/Cas 9 toolkit for multiplex genome editing in plants. BMC Plant Biol, 14:327.
doi: 10.1186/s12870-014-0327-y URL |
| [43] | Yang Meng-xia. 2020. Creation of tomato male sterile material by gene editing[M. D. Dissertation]. Beijing: Chinese Academy of Agricultural Sciences. (in Chinese) |
| 杨孟霞. 2020. 利用基因编辑技术创制番茄雄性不育材料的研究[硕士论文]. 北京: 中国农业科学院. | |
| [44] |
Ye J, Wang X, Hu T, Zhang F, Wang B, Li C, Yang T, Li H, Lu Y, Giovannoni J J, Zhang Y, Ye Z. 2017. An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell, 29 (9):2249-2268.
doi: 10.1105/tpc.17.00211 URL |
| [45] |
Yu W, Wang L, Zhao R, Sheng J, Zhang S, Li R, Shen L. 2019. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol, 19 (1):354.
doi: 10.1186/s12870-019-1939-z |
| [46] |
Zhang Aiping, Liu Jiangna, Yan Jianjun, Zhang Xiying, Bai Yunfeng. 2022. Progress and prospect of genome editing in tomato. Acta Horticulturae Sinica, 49 (1):221-232. (in Chinese)
doi: 10.16420/j.issn.0513-353x.2020-0976 URL |
|
张爱萍, 刘江娜, 闫建俊, 张西英, 白云凤. 2022. 番茄基因编辑研究进展和前景. 园艺学报, 49 (1):221-232.
doi: 10.16420/j.issn.0513-353x.2020-0976 URL |
|
| [47] |
Zhang Z, Mao Y, Ha S, Liu W, Botella J R, Zhu J K. 2016. A multiplex CRISPR/Cas 9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep, 35 (7):1519-1533.
doi: 10.1007/s00299-015-1900-z URL |
| [48] |
Zhou Xiang-chun, Xing Yong-zhong. 2016. The application of genome editing in identification of plant gene function and crop breeding. Hereditas(Beijing), 38 (3):227-242. (in Chinese)
doi: 10.16288/j.yczz.15-327 pmid: 27001477 |
| 周想春, 邢永忠. 2016. 基因组编辑技术在植物基因功能鉴定及作物育种中的应用. 遗传, 38 (3):227-242. | |
| [49] |
Zsögön A, Čermák T, Naves E R, Notini M M, Edel K H, Weinl S, Freschi L, Voytas D F, Kudla J, Peres L E P. 2018. De novo domestication of wild tomato using genome editing. Nat Biotechnol, 36 (12):1211-1216.
doi: 10.1038/nbt.4272 |
| [1] | 苏银玲, 杨子祥, 但 忠, 麻继仙, 杨 龙, 李易蓉, 唐正富, 汪玲敏, 木万福. 番茄新品种‘云番茄 68’[J]. 园艺学报, 2023, 50(S1): 63-64. |
| [2] | 樊 娟, 沈松真, 苗青青, 张乐辉, 姚恩鹏, 裴卓强. 番茄新品种‘卫红 16 号’[J]. 园艺学报, 2023, 50(S1): 65-66. |
| [3] | 张辉, 朱为民, 朱龙英, 杨学东, 张迎迎, 田守波, 万延慧, 刘雅慧, 杨志杰. 樱桃番茄新品种‘沪樱9号’[J]. 园艺学报, 2023, 50(6): 1379-1380. |
| [4] | 李国斌, 蔡梁玉, 肖立成, 王家发, 张得迪, 张俊红. 利用CRISPR/Cas9技术创制番茄粉果和无绿肩材料[J]. 园艺学报, 2023, 50(5): 985-999. |
| [5] | 唐海霞, 裴广营, 张琼, 王中堂. 枣果实相关性状QTL定位分析[J]. 园艺学报, 2023, 50(4): 754-764. |
| [6] | 徐悦, 李子雄, 陈婕, 孙亮. 2,4-D调控番茄果形的转录基础[J]. 园艺学报, 2023, 50(4): 802-814. |
| [7] | 郑锦荣, 聂俊, 李艳红, 谭德龙, 谢玉明, 张长远. 樱桃番茄新品种‘粤科达101’[J]. 园艺学报, 2023, 50(4): 909-910. |
| [8] | 刘玉菡, 陶宁, 王庆国, 李清清. 番茄中ABC转运蛋白SlABCG23调控茉莉酸信号途径[J]. 园艺学报, 2023, 50(3): 559-568. |
| [9] | 史洪丽, 李腊, 郭翠梅, 余婷婷, 简伟, 杨星勇. 番茄灰霉病生防菌株TL1的分离、鉴定及其生防能力分析[J]. 园艺学报, 2023, 50(1): 79-90. |
| [10] | 忽靖宇, 阙开娟, 缪田丽, 吴少政, 王田田, 张磊, 董鲜, 季鹏章, 董家红. 侵染鸢尾的番茄斑萎病毒鉴定[J]. 园艺学报, 2023, 50(1): 170-176. |
| [11] | 郑积荣, 王同林, 胡松申. 高品质番茄新品种‘杭杂603’[J]. 园艺学报, 2022, 49(S2): 103-104. |
| [12] | 郑积荣, 王同林. 番茄新品种‘杭杂601’[J]. 园艺学报, 2022, 49(S2): 105-106. |
| [13] | 郑积荣, 王同林. 樱桃番茄新品种‘杭杂503’[J]. 园艺学报, 2022, 49(S2): 107-108. |
| [14] | 黄婷婷, 刘淑芹, 张永志, 李 平, 张志焕, 宋立波. 樱桃番茄新品种‘樱莎红4号’[J]. 园艺学报, 2022, 49(S2): 109-110. |
| [15] | 张前荣, 李大忠, 裘波音, 林 珲, 马慧斐, 叶新如, 刘建汀, 朱海生, 温庆放. 设施番茄新品种‘闽农科2号’[J]. 园艺学报, 2022, 49(S1): 73-74. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司