
园艺学报 ›› 2022, Vol. 49 ›› Issue (4): 778-790.doi: 10.16420/j.issn.0513-353x.2021-0347
杨易1, 黎庭耀1, 李国景2, 陈汉才1, 沈卓1, 周轩1, 吴增祥1, 吴新义2, 张艳1,*( )
)
收稿日期:2021-11-01
修回日期:2022-01-18
出版日期:2022-04-25
发布日期:2022-04-24
通讯作者:
张艳
E-mail:893251473@qq.com
基金资助:
YANG Yi1, LI Tingyao1, LI Guojing2, CHEN Hancai1, SHEN Zhuo1, ZHOU Xuan1, WU Zengxiang1, WU Xinyi2, ZHANG Yan1,*( )
)
Received:2021-11-01
Revised:2022-01-18
Online:2022-04-25
Published:2022-04-24
Contact:
ZHANG Yan
E-mail:893251473@qq.com
摘要:
选取3份长豇豆种质资源进行全基因组重测序,利用重测序数据挖掘InDel位点,根据这些位点开发分子标记,并验证这些标记的有效性和应用价值。经过严格筛选,获得981个易于凝胶电泳检测的长度大于30 bp且多态性高的InDel位点,挑选其中在基因组上均匀分布的165个设计引物并进行验证。用6份长豇豆材料对165对引物进行初筛,其中162对可获得PCR扩增产物,共扩增出673条条带;85对引物表现出多态性,多态性带333条(占总带数的49.48%)。利用85个多态性高的标记对173份长豇豆资源进行基因分型和遗传多样性分析,共检测到333个等位位点;多态性信息含量(PIC)为0.01 ~ 0.37,平均值为0.29;基因多样性指数变异范围为0.01 ~ 0.50,平均值为0.36,表明现有长豇豆资源遗传背景相对狭窄。通过聚类分析,173份长豇豆被分为2个类群,成功将不同类型长豇豆划分到不同亚群。另外,选用亲本间多态性标记用于6个杂交组合的F1代真实性鉴定,鉴定出5个真实杂交种。本研究所开发的多态性 InDel 标记在长豇豆遗传多样性分析、杂交种纯度鉴定、遗传连锁图谱构建、基因定位和分子标记辅助选择育种等方面具有很好的应用前景。
中图分类号:
杨易, 黎庭耀, 李国景, 陈汉才, 沈卓, 周轩, 吴增祥, 吴新义, 张艳. 基于重测序的长豇豆基因组InDel标记开发及应用[J]. 园艺学报, 2022, 49(4): 778-790.
YANG Yi, LI Tingyao, LI Guojing, CHEN Hancai, SHEN Zhuo, ZHOU Xuan, WU Zengxiang, WU Xinyi, ZHANG Yan. Development and Application of Insertion-Deletion(InDel)Markers in Asparagus Bean Based on Whole Genome Re-sequencing Data[J]. Acta Horticulturae Sinica, 2022, 49(4): 778-790.
| 编号No. | 分类Type | 资源数量Number | 比例/%Ratio | 命名Name |
|---|---|---|---|---|
| 1 | 油白类White type | 68 | 39.31 | 1-1,……1-68 |
| 2 | 油青类Green type | 45 | 26.01 | 2-1,……2-45 |
| 3 | 紫豇豆Purple type | 16 | 9.25 | 3-1,……3-16 |
| 4 | 特青类Dark green type | 10 | 5.78 | 4-1,……4-10 |
| 5 | 纯白类Pure white type | 14 | 8.09 | 5-1,……5-14 |
| 6 | 灰青类Gray green type | 7 | 4.05 | 6-1,……6-7 |
| 7 | 无架豆类(矮生型)Shelf free type | 4 | 2.31 | 7-1,……7-4 |
| 8 | 密豆类(籽粒密度高) Pod with high density grains type | 9 | 5.20 | 8-1,……8-9 |
表1 173份长豇豆种质资源的分类编号及数量
Table 1 The classification numbers of 173 asparagus bean germplasm resources
| 编号No. | 分类Type | 资源数量Number | 比例/%Ratio | 命名Name |
|---|---|---|---|---|
| 1 | 油白类White type | 68 | 39.31 | 1-1,……1-68 |
| 2 | 油青类Green type | 45 | 26.01 | 2-1,……2-45 |
| 3 | 紫豇豆Purple type | 16 | 9.25 | 3-1,……3-16 |
| 4 | 特青类Dark green type | 10 | 5.78 | 4-1,……4-10 |
| 5 | 纯白类Pure white type | 14 | 8.09 | 5-1,……5-14 |
| 6 | 灰青类Gray green type | 7 | 4.05 | 6-1,……6-7 |
| 7 | 无架豆类(矮生型)Shelf free type | 4 | 2.31 | 7-1,……7-4 |
| 8 | 密豆类(籽粒密度高) Pod with high density grains type | 9 | 5.20 | 8-1,……8-9 |

图1 部分InDel引物在6份豇豆材料的扩增结果 M:100 ~ 600 bp的DNA marker。点样顺序从左到右分别为豇豆材料1-8、1-17、2-22、2-8、2-10、5-12,图中数字代表引物序号。
Fig. 1 Amplification results of some InDel primers in six asparagus bean accessions M:100-600 bp DNA marker. The corresponding bands of asparagus bean materials are 1-8,1-17,2-22,2-8,2-10 and 5-12 from left to right,the numbers in the figure represent primer serial number.
| 染色体编号 Chromosome number | 引物数 Number of primer | 有效扩增 引物数 Effective amplification primer | 有效引物 含量/% Effective amplification primer content | 多态引物 Polymorphic primers number | 多态引物 含量/% Polymorphic primers content | 扩增总条数 Total number of amplified band | 多态条带数 Number of polymorphic band | 多态条带 比例/% Ratio |
|---|---|---|---|---|---|---|---|---|
| Vu1 | 17 | 17 | 100.00 | 7 | 41.18 | 94 | 35 | 37.23 |
| Vu 2 | 13 | 13 | 100.00 | 6 | 46.15 | 60 | 24 | 40.00 |
| Vu 3 | 22 | 22 | 100.00 | 7 | 31.82 | 95 | 19 | 20.00 |
| Vu 4 | 19 | 19 | 100.00 | 13 | 68.42 | 76 | 47 | 61.84 |
| Vu 5 | 14 | 14 | 100.00 | 10 | 71.43 | 48 | 37 | 77.08 |
| Vu 6 | 17 | 16 | 94.12 | 10 | 58.82 | 54 | 38 | 70.37 |
| Vu 7 | 9 | 8 | 88.89 | 5 | 55.56 | 27 | 22 | 81.48 |
| Vu 8 | 11 | 10 | 90.91 | 4 | 36.36 | 45 | 14 | 31.11 |
| Vu 9 | 16 | 16 | 100.00 | 10 | 62.50 | 66 | 43 | 65.15 |
| Vu 10 | 16 | 16 | 100.00 | 9 | 56.25 | 61 | 42 | 68.85 |
| Vu 11 | 11 | 11 | 100.00 | 4 | 36.36 | 47 | 12 | 25.53 |
| 总数Total | 165 | 162 | 98.18 | 85 | 51.52 | 673 | 333 | 49.48 |
表2 InDel标记引物扩增结果数据统计
Table 2 The statistics of InDel marker primers amplification results
| 染色体编号 Chromosome number | 引物数 Number of primer | 有效扩增 引物数 Effective amplification primer | 有效引物 含量/% Effective amplification primer content | 多态引物 Polymorphic primers number | 多态引物 含量/% Polymorphic primers content | 扩增总条数 Total number of amplified band | 多态条带数 Number of polymorphic band | 多态条带 比例/% Ratio |
|---|---|---|---|---|---|---|---|---|
| Vu1 | 17 | 17 | 100.00 | 7 | 41.18 | 94 | 35 | 37.23 |
| Vu 2 | 13 | 13 | 100.00 | 6 | 46.15 | 60 | 24 | 40.00 |
| Vu 3 | 22 | 22 | 100.00 | 7 | 31.82 | 95 | 19 | 20.00 |
| Vu 4 | 19 | 19 | 100.00 | 13 | 68.42 | 76 | 47 | 61.84 |
| Vu 5 | 14 | 14 | 100.00 | 10 | 71.43 | 48 | 37 | 77.08 |
| Vu 6 | 17 | 16 | 94.12 | 10 | 58.82 | 54 | 38 | 70.37 |
| Vu 7 | 9 | 8 | 88.89 | 5 | 55.56 | 27 | 22 | 81.48 |
| Vu 8 | 11 | 10 | 90.91 | 4 | 36.36 | 45 | 14 | 31.11 |
| Vu 9 | 16 | 16 | 100.00 | 10 | 62.50 | 66 | 43 | 65.15 |
| Vu 10 | 16 | 16 | 100.00 | 9 | 56.25 | 61 | 42 | 68.85 |
| Vu 11 | 11 | 11 | 100.00 | 4 | 36.36 | 47 | 12 | 25.53 |
| 总数Total | 165 | 162 | 98.18 | 85 | 51.52 | 673 | 333 | 49.48 |

图2 引物Vu6-94在173份长豇豆材料的扩增图 M:100 ~ 600 bp的DNA marker,“↑”指示材料2-23。
Fig. 2 Amplification products of marker Vu6-94 in 173 asparagus bean accessions M:100-600 bp DNA markers,“↑”indicating asparagus bean accession 2-23.
| 上游引物 Forward primer | 物理位置Physical location | 引物序列(5′-3′) Forward primer sequence | 下游引物 Reverse primer | 物理位置 Physical location | 引物序列(5′-3′) Reverse primer sequence |
|---|---|---|---|---|---|
| Vu1-4f | 9 635 438 | CTTGTGCATTGTTCTTGGTTCA | Vu1-4r | 9 635 460 | AGGGTAACTTGTTTTCAGCCAT |
| Vu1-5f | 12 339 675 | ATGGATAGGAGGGGAGGAAATT | Vu1-5r | 12 339 697 | TTTAGTCCCCTCTCCTCTACAC |
| Vu1-6f | 14 580 618 | TCGTTCTTTTACCATCTTCGCA | Vu1-6r | 14 580 640 | TTGAAGCGTGTTTAAATTACTGA |
| Vu1-10f | 23 979 363 | GAGTGATAGATCTTGGAGGTGC | Vu1-10r | 23 979 385 | CCTTCATAACCAGCTAAAAGGC |
| Vu1-12f | 26 295 823 | GCCCAACCGTGTTAAGTCTAAT | Vu1-12r | 26 295 845 | GAGGTTTCTAAGCACATGAGGT |
| Vu1-13f | 28 949 546 | ACATGCTGCATTCTCTACCATT | Vu1-13r | 28 949 568 | TCACGACGAGAACCATATTTGA |
| Vu1-17f | 41 148 093 | AGAAATTGGAGGAAAGGTGAGC | Vu1-17r | 41 148 115 | GACGATGATGGTGATGATGACA |
| Vu2-19f | 18 161 417 | AAGGTGGCGTGAATAATGTCTT | Vu2-19r | 18 161 439 | TTATCCAGAACTCCTTCAAGCC |
| Vu2-20f | 19 179 823 | ATCTTCATTGTCCATTCCTGCA | Vu2-20r | 19 179 845 | TTGGGTTGATCTTGTATGCCAA |
| Vu2-22f | 22 321 069 | CGTGAAGCTCAATCGAATCTTT | Vu2-22r | 22 321 091 | TCCAATCCATCATTCATTACGC |
| Vu2-25f | 26 891 508 | AAAGCATGTCATTAGCAGCATC | Vu2-25r | 26 891 530 | TGAAGAGCTTGAACGTGAGAAT |
| Vu2-28f | 30 986 485 | TTTCACGTTGGATCAAGCAAAC | Vu2-28r | 30 986 507 | TGTCACCAGAAAAGGCCATAAT |
| Vu2-29f | 32 863 650 | TTAGATTGGACAGTGAAGCTCG | Vu2-29r | 32 863 672 | TGATGCTGTTTGGATTGTGGG |
| Vu3-32f | 5 016 424 | TTCGTTTCAAACCTCCACTTCT | Vu3-32r | 5 016 446 | CTGTCTACCTGCAGTCGATTAG |
| Vu3-37f | 23 098 591 | CATGTCCACCCTGAAGCTATAG | Vu3-37r | 23 098 613 | AGAAAAGCAATACAAACGTGGC |
| Vu3-38f | 25 801 706 | CTGCTCTCAACCTTATCCCAAA | Vu3-38r | 25 801 728 | CTCTTGCAGTCCCTATTGTCAA |
| Vu3-44f | 39 770 758 | ATTTCAATAGTTCTGGCGAGCA | Vu3-44r | 39 770 780 | CACAAAGTGGCATATCTGGTTT |
| Vu3-45f | 42 848 262 | TATCCATGGGTTCAAATCGGAG | Vu3-45r | 42 848 284 | AGAACTGGCGAGAAGAAATGAT |
| Vu3-46f | 43 685 697 | CAGGCTTTGGTCATTGGATTTT | Vu3-46r | 43 685 719 | AAGAAAGAGACCTGAAGTGCTC |
| Vu3-52f | 65 121 995 | ATTGGGTGTGACATTCTTTACG | Vu3-52r | 65 122 017 | AACCTAATCGCACTGAACCGS |
| Vu4-53f | 4 405 112 | TGCAGCATTGAAGAATCTCCAA | Vu4-53r | 4 405 134 | GTGCAAGAATATGTGTTCCAGC |
| Vu4-54f | 4 703 051 | CTGGAATTGCAGTCATCTATCCT | Vu4-54r | 4 703 074 | AATTCCATTCCCGTGTTCTGAA |
| Vu4-55f | 7 730 103 | GGATAGCCAAAACCATAGTAGCT | Vu4-55r | 7 730 126 | CAAACTGTGGTGGTGAATATGA |
| Vu4-56f | 9 241 355 | GTGGTTATCTTGCTCTAGGTCC | Vu4-56r | 9 241 377 | TCTAAAGGGCACGATCAACATT |
| Vu4-57f | 11 119 748 | CTCATATATTTGTAGGGCCGGG | Vu4-57r | 11 119 770 | TGGGACCTTCAAGTGCAATTAA |
| Vu4-59f | 14 278 136 | AGGTCAAGTACATGTTGCATGA | Vu4-59r | 14 278 158 | GCACTGTTTTGGTTCTTAGCAT |
| Vu4-60f | 15 050 272 | GGGAGTCATTGCTGTTAGAGTT | Vu4-60r | 15 050 294 | GAAAAGGGTATGCATGAACAAC |
| Vu4-62f | 16 066 798 | ATTTTGGAGTGTATCAGTGGGG | Vu4-62r | 16 066 820 | ACTGAAAATCGCACTGGAAAAC |
| Vu4-63f | 16 457 840 | GGGTCCTTTTCTATTGCTCAGT | Vu4-63r | 16 457 862 | TCCTGAAACTGTCTTTCCCATC |
| Vu4-64f | 18 682 066 | GGGAACAAAAGATGCAAGTGAG | Vu4-64r | 18 682 088 | CAAAATTTGGGTTGTAGTCAGC |
| Vu4-65f | 19 635 565 | TTAACAAACCCATGAAACAGGC | Vu4-65r | 19 635 587 | GATATGGTTCTGTTGGTGGGAG |
| Vu4-70f | 37 023 441 | GGAGTTGAGACAGTAACATGGA | Vu4-70r | 37 023 463 | GGGTTTGATTCACTTTTGTCCC |
| Vu4-71f | 40 172 227 | TCACAACAGACAAACTCTACGT | Vu4-71r | 40 172 249 | GCATGCCACCATAATCACAAAT |
| Vu5-72f | 879 582 | GCCCCTCGATATTCTCAGATTC | Vu5-72r | 879 604 | AGTTTTGGCTAGCAGGTATTGT |
| Vu5-73f | 2 307 937 | TGTTCTTATGCATGGGTCAGTT | Vu5-73r | 2 307 959 | GCTCAAACAAGTGCAAGATCAT |
| Vu5-74f | 4 191 243 | CGAAGAAAGACAGTTGTTTGCA | Vu5-74r | 4 191 265 | CTGTTGATAGAGTGTGAGTCCC |
| Vu5-77f | 19 695 657 | CTTCACTGCTCTCCTTGCTATT | Vu5-77r | 19 695 679 | CTTGAAGTTGTGAGTGGACAGA |
| Vu5-78f | 24 528 035 | TCCCTGAGCATCCATTATTTCC | Vu5-78r | 24 528 057 | GTCATTCGGTCCATTTCACAAG |
| Vu5-80f | 31 810 455 | GTTGAAAGTGTATTGTTCTGCG | Vu5-80r | 31 810 477 | GGGACCACAGTAAAATCATGAC |
| Vu5-81f | 34 174 933 | ACATCGTGGTGCTCTTTCATTA | Vu5-81r | 34 174 955 | TGACTGGAAACCTTCTAACTGG |
| Vu5-82f | 41 723 563 | AGTTATTGGAGAACATGCCTCT | Vu5-82r | 41 723 585 | GGCGTTTAAGATTTAGTCGACA |
| Vu5-84f | 46 562 084 | CGGACAAAAGAAATAACCTGCA | Vu5-84r | 46 562 106 | TGTTCTCCAATAGTTCCTCACG |
| Vu5-85f | 48 539 129 | ACTTTCACCACTTACTCTCACT | Vu5-85r | 48 539 151 | CACATGTAAACAAGTAGTAGACGC |
| Vu6-87f | 2 458 695 | CCTACCCAGCCAATTTTGTCTA | Vu6-87r | 2 458 717 | CTGAATTCTCCAGGACTGATTC |
| Vu6-88f | 4 157 381 | CTGTACCAATCCACATGAACCT | Vu6-88r | 4 157 403 | CTGAATTGAACGAAGAAGTGCG |
| Vu6-90f | 13 739 218 | GCTTCCTAAGATTGCAACCTTG | Vu6-90r | 13 739 240 | AAACTCTAAGACCCAACCTGTC |
| Vu6-94f | 19 440 325 | AAGGTTTAGTTTAGGCTGTCCC | Vu6-94r | 19 440 347 | TGGAAAATGTGCACAACTAGGA |
| Vu6-95f | 20 702 486 | TTGTGTATTGGGTAAGTGCCAA | Vu6-95r | 20 702 508 | TCTTCCTCAGTCCCAATACTCA |
| Vu6-97f | 23 626 647 | ATTTTCTTCTTGGCTGCTGTTC | Vu6-97r | 23 626 669 | CTCTCTCCCACTCCTTTCAATG |
| Vu6-99f | 28 111 300 | CACTCCCAAGGAAGATGAGATC | Vu6-99r | 28 111 322 | GTTCTCAGGGGAAAAGGTGATT |
| Vu6-100f | 30 559 787 | ACATCTATCGCCAATCTCACAG | Vu6-100r | 30 559 809 | CAAATGAAGTGGGGAGGAAGAA |
| Vu6-101f | 31 914 466 | CACTTGAAGACGCTAAGAACAC | Vu6-101r | 31 914 488 | ACCTCCAACAAGTTCATGATCA |
| Vu6-102f | 32 539 051 | TATTGGAGAAAGGGCATAGCAG | Vu6-102r | 32 539 073 | CTTTTCACCGGTATCCCTTCTT |
| Vu7-105f | 13 121 356 | TCCTCTACCTTTCACCAAGCTA | Vu7-105r | 13 121 378 | ATGGTATGTCTTGTGATGGCAA |
| Vu7-106f | 23 354 328 | GATAGCTCACAACACAGACACT | Vu7-106r | 23 354 350 | TGTATCGGTACTCACATGCTTC |
| Vu7-108f | 29 923 695 | CACTTAGCATCTGATGGCCTTA | Vu7-108r | 29 923 717 | ACTGGGGTATTTCAAAGTGAGG |
| Vu7-109f | 31 969 597 | AAAGGGGTCACTCATACACATG | Vu7-109r | 31 969 619 | GACACCACTTTAACCCAAAACC |
| Vu7-110f | 35 390 277 | TGTGGTAGCTCAGTGTTAGTTG | Vu7-110r | 35 390 299 | AATATAACATAGGCCGGCTGAC |
| Vu8-113f | 6 256 060 | TGCGTGTAAGTGTCAGAGAAAT | Vu8-113r | 6 256 082 | AAGTGTGGTGAGGAAAGGTATG |
| Vu8-115f | 10 801 610 | TTGCTTAACCTGAGATGAACGT | Vu8-115r | 10 801 632 | ACGGGAGGTTATAGGCTATTTG |
| Vu8-116f | 21 415 000 | GACACAAGATCGAATCCAAGGA | Vu8-116r | 21 415 022 | GCACTGCATCTTTCACTTTCAT |
| Vu9-122f | 1 063 922 | CCTGGTACTTGGACTCAGATTT | Vu9-122r | 1 063 944 | CAGCTGTTGATGTTGAACCAAA |
| Vu9-123f | 2 491 158 | TTGTGTTAAAGGTGGTGCATTG | Vu9-123r | 2 491 180 | AGGGGTAAAGCAAGCAACATAT |
| Vu9-124f | 4 327 558 | GTCTCTATGGCGGTCAAGTTTA | Vu9-124r | 4 327 580 | CTTCATGATCTATCTGCCCCAG |
| Vu9-125f | 5 918 421 | GTATCGGATGCTGTTATTTGGG | Vu9-125r | 5 918 443 | GTGGAATTTCTATTTTGCCTGC |
| Vu9-126f | 7 169 197 | GATCATTGAACTCCCTTGGTCA | Vu9-126r | 7 169 219 | GTTAGCAACAGCAACCGATAAG |
| Vu9-127f | 9 039 864 | GCAGTGACCTCTCAATTCATCT | Vu9-127r | 9 039 886 | CCCTGCATCTCCATTTTGATTT |
| Vu9-129f | 15 935 910 | ACGATTCTACTGCTTAGCCATC | Vu9-129r | 15 935 932 | GCGAGAATTAAAGGAAACGTCA |
| Vu9-131f | 22 071 671 | CCCCGAATAAGTGTTCTAGGTG | Vu9-131r | 22 071 693 | CAAAACATGGCCATACATCTCG |
| Vu9-135f | 33 519 683 | CACTGCAACAATTATACGCCTT | Vu9-135r | 33 519 705 | TGTCACAGCTCCAAAGAATGAT |
| Vu9-136f | 34 865 479 | AAAGTTGTAGGCTAAGGTGGAC | Vu9-136r | 34 865 501 | CACAGATAACGAGGAGATAGGC |
| Vu9-138f | 40 694 878 | AAACCCACTCATGTACAAGACT | Vu9-138r | 40 694 900 | TGGTGTTCATCAAGATTTCACG |
| Vu10-141f | 5 081 721 | GCGATTGATTTAAGGCACAGTA | Vu10-141r | 5 081 743 | AAGTCAGGGGTTTTGATCAAGT |
| Vu10-142f | 7 985 569 | AGTCACAGTTTCAGTCACCAAA | Vu10-142r | 7 985 591 | ACGGAGATCAGTCACTATAACC |
| Vu10-144f | 12 679 952 | AGGGCTTCAATTTTCTCCTTCA | Vu10-144r | 12 679 974 | GAATTTGGAAGGCTGTTGTTGT |
| Vu10-145f | 15 994 371 | CCGGAAAACTCACAACATCATT | Vu10-145r | 15 994 393 | CCAACTTTTGAATGCTTGACCA |
| Vu10-146f | 17 013 915 | ACTGGTCTTCATTTTGTGCTGA | Vu10-146r | 17 013 937 | AGGCTTCATTGGGTAGTGTTAC |
| Vu10-147f | 23 281 009 | ACTTCATCAATGTCCACCTCTT | Vu10-147r | 23 281 031 | CGCAACGGATAGGAAATGGATA |
| Vu10-149f | 28 197 383 | TGTTTCTTGCGCTGACATAGTA | Vu10-149r | 28 197 405 | TTAGCAGTTGTCGAGTCTCTTC |
| Vu10-152f | 36 129 524 | AGTAAGCACAGTCTTAGAGCAA | Vu10-152r | 36 129 546 | TAGGCAATTCACAGTCTCTTGG |
| Vu10-154f | 39 650 768 | GACAGAAGAGGGGTTCAGAATC | Vu10-154r | 39 650 790 | AGTTCAAGTACATGCTCCAACA |
| Vu11-157f | 10 620 552 | CATTTCGCTCCCTGAAGTTTC | Vu11-157r | 10 620 573 | TGGTTTAGAAAGAGGTTACGTG |
| Vu11-160f | 24 401 745 | CCAACACGCTTTCATGATTCAA | Vu11-160r | 24 401 767 | GGTCTCGTGCAAAAGTAATGTC |
| Vu11-161f | 28 839 447 | AGTATCCAACAACCAACCATCA | Vu11-161r | 28 839 469 | GCGTGGAAATATGTATCGATCG |
| Vu11-162f | 32 266 203 | GGTCACGTCCAGAGATAATTACT | Vu11-162r | 32 266 226 | CACTTGCCAATATTCACAGCA |
表3 85对长豇豆InDel标记序列位置信息及对应引物序列
Table 3 Location and corresponding sequences information of 85 pairs of asparagus bean primers
| 上游引物 Forward primer | 物理位置Physical location | 引物序列(5′-3′) Forward primer sequence | 下游引物 Reverse primer | 物理位置 Physical location | 引物序列(5′-3′) Reverse primer sequence |
|---|---|---|---|---|---|
| Vu1-4f | 9 635 438 | CTTGTGCATTGTTCTTGGTTCA | Vu1-4r | 9 635 460 | AGGGTAACTTGTTTTCAGCCAT |
| Vu1-5f | 12 339 675 | ATGGATAGGAGGGGAGGAAATT | Vu1-5r | 12 339 697 | TTTAGTCCCCTCTCCTCTACAC |
| Vu1-6f | 14 580 618 | TCGTTCTTTTACCATCTTCGCA | Vu1-6r | 14 580 640 | TTGAAGCGTGTTTAAATTACTGA |
| Vu1-10f | 23 979 363 | GAGTGATAGATCTTGGAGGTGC | Vu1-10r | 23 979 385 | CCTTCATAACCAGCTAAAAGGC |
| Vu1-12f | 26 295 823 | GCCCAACCGTGTTAAGTCTAAT | Vu1-12r | 26 295 845 | GAGGTTTCTAAGCACATGAGGT |
| Vu1-13f | 28 949 546 | ACATGCTGCATTCTCTACCATT | Vu1-13r | 28 949 568 | TCACGACGAGAACCATATTTGA |
| Vu1-17f | 41 148 093 | AGAAATTGGAGGAAAGGTGAGC | Vu1-17r | 41 148 115 | GACGATGATGGTGATGATGACA |
| Vu2-19f | 18 161 417 | AAGGTGGCGTGAATAATGTCTT | Vu2-19r | 18 161 439 | TTATCCAGAACTCCTTCAAGCC |
| Vu2-20f | 19 179 823 | ATCTTCATTGTCCATTCCTGCA | Vu2-20r | 19 179 845 | TTGGGTTGATCTTGTATGCCAA |
| Vu2-22f | 22 321 069 | CGTGAAGCTCAATCGAATCTTT | Vu2-22r | 22 321 091 | TCCAATCCATCATTCATTACGC |
| Vu2-25f | 26 891 508 | AAAGCATGTCATTAGCAGCATC | Vu2-25r | 26 891 530 | TGAAGAGCTTGAACGTGAGAAT |
| Vu2-28f | 30 986 485 | TTTCACGTTGGATCAAGCAAAC | Vu2-28r | 30 986 507 | TGTCACCAGAAAAGGCCATAAT |
| Vu2-29f | 32 863 650 | TTAGATTGGACAGTGAAGCTCG | Vu2-29r | 32 863 672 | TGATGCTGTTTGGATTGTGGG |
| Vu3-32f | 5 016 424 | TTCGTTTCAAACCTCCACTTCT | Vu3-32r | 5 016 446 | CTGTCTACCTGCAGTCGATTAG |
| Vu3-37f | 23 098 591 | CATGTCCACCCTGAAGCTATAG | Vu3-37r | 23 098 613 | AGAAAAGCAATACAAACGTGGC |
| Vu3-38f | 25 801 706 | CTGCTCTCAACCTTATCCCAAA | Vu3-38r | 25 801 728 | CTCTTGCAGTCCCTATTGTCAA |
| Vu3-44f | 39 770 758 | ATTTCAATAGTTCTGGCGAGCA | Vu3-44r | 39 770 780 | CACAAAGTGGCATATCTGGTTT |
| Vu3-45f | 42 848 262 | TATCCATGGGTTCAAATCGGAG | Vu3-45r | 42 848 284 | AGAACTGGCGAGAAGAAATGAT |
| Vu3-46f | 43 685 697 | CAGGCTTTGGTCATTGGATTTT | Vu3-46r | 43 685 719 | AAGAAAGAGACCTGAAGTGCTC |
| Vu3-52f | 65 121 995 | ATTGGGTGTGACATTCTTTACG | Vu3-52r | 65 122 017 | AACCTAATCGCACTGAACCGS |
| Vu4-53f | 4 405 112 | TGCAGCATTGAAGAATCTCCAA | Vu4-53r | 4 405 134 | GTGCAAGAATATGTGTTCCAGC |
| Vu4-54f | 4 703 051 | CTGGAATTGCAGTCATCTATCCT | Vu4-54r | 4 703 074 | AATTCCATTCCCGTGTTCTGAA |
| Vu4-55f | 7 730 103 | GGATAGCCAAAACCATAGTAGCT | Vu4-55r | 7 730 126 | CAAACTGTGGTGGTGAATATGA |
| Vu4-56f | 9 241 355 | GTGGTTATCTTGCTCTAGGTCC | Vu4-56r | 9 241 377 | TCTAAAGGGCACGATCAACATT |
| Vu4-57f | 11 119 748 | CTCATATATTTGTAGGGCCGGG | Vu4-57r | 11 119 770 | TGGGACCTTCAAGTGCAATTAA |
| Vu4-59f | 14 278 136 | AGGTCAAGTACATGTTGCATGA | Vu4-59r | 14 278 158 | GCACTGTTTTGGTTCTTAGCAT |
| Vu4-60f | 15 050 272 | GGGAGTCATTGCTGTTAGAGTT | Vu4-60r | 15 050 294 | GAAAAGGGTATGCATGAACAAC |
| Vu4-62f | 16 066 798 | ATTTTGGAGTGTATCAGTGGGG | Vu4-62r | 16 066 820 | ACTGAAAATCGCACTGGAAAAC |
| Vu4-63f | 16 457 840 | GGGTCCTTTTCTATTGCTCAGT | Vu4-63r | 16 457 862 | TCCTGAAACTGTCTTTCCCATC |
| Vu4-64f | 18 682 066 | GGGAACAAAAGATGCAAGTGAG | Vu4-64r | 18 682 088 | CAAAATTTGGGTTGTAGTCAGC |
| Vu4-65f | 19 635 565 | TTAACAAACCCATGAAACAGGC | Vu4-65r | 19 635 587 | GATATGGTTCTGTTGGTGGGAG |
| Vu4-70f | 37 023 441 | GGAGTTGAGACAGTAACATGGA | Vu4-70r | 37 023 463 | GGGTTTGATTCACTTTTGTCCC |
| Vu4-71f | 40 172 227 | TCACAACAGACAAACTCTACGT | Vu4-71r | 40 172 249 | GCATGCCACCATAATCACAAAT |
| Vu5-72f | 879 582 | GCCCCTCGATATTCTCAGATTC | Vu5-72r | 879 604 | AGTTTTGGCTAGCAGGTATTGT |
| Vu5-73f | 2 307 937 | TGTTCTTATGCATGGGTCAGTT | Vu5-73r | 2 307 959 | GCTCAAACAAGTGCAAGATCAT |
| Vu5-74f | 4 191 243 | CGAAGAAAGACAGTTGTTTGCA | Vu5-74r | 4 191 265 | CTGTTGATAGAGTGTGAGTCCC |
| Vu5-77f | 19 695 657 | CTTCACTGCTCTCCTTGCTATT | Vu5-77r | 19 695 679 | CTTGAAGTTGTGAGTGGACAGA |
| Vu5-78f | 24 528 035 | TCCCTGAGCATCCATTATTTCC | Vu5-78r | 24 528 057 | GTCATTCGGTCCATTTCACAAG |
| Vu5-80f | 31 810 455 | GTTGAAAGTGTATTGTTCTGCG | Vu5-80r | 31 810 477 | GGGACCACAGTAAAATCATGAC |
| Vu5-81f | 34 174 933 | ACATCGTGGTGCTCTTTCATTA | Vu5-81r | 34 174 955 | TGACTGGAAACCTTCTAACTGG |
| Vu5-82f | 41 723 563 | AGTTATTGGAGAACATGCCTCT | Vu5-82r | 41 723 585 | GGCGTTTAAGATTTAGTCGACA |
| Vu5-84f | 46 562 084 | CGGACAAAAGAAATAACCTGCA | Vu5-84r | 46 562 106 | TGTTCTCCAATAGTTCCTCACG |
| Vu5-85f | 48 539 129 | ACTTTCACCACTTACTCTCACT | Vu5-85r | 48 539 151 | CACATGTAAACAAGTAGTAGACGC |
| Vu6-87f | 2 458 695 | CCTACCCAGCCAATTTTGTCTA | Vu6-87r | 2 458 717 | CTGAATTCTCCAGGACTGATTC |
| Vu6-88f | 4 157 381 | CTGTACCAATCCACATGAACCT | Vu6-88r | 4 157 403 | CTGAATTGAACGAAGAAGTGCG |
| Vu6-90f | 13 739 218 | GCTTCCTAAGATTGCAACCTTG | Vu6-90r | 13 739 240 | AAACTCTAAGACCCAACCTGTC |
| Vu6-94f | 19 440 325 | AAGGTTTAGTTTAGGCTGTCCC | Vu6-94r | 19 440 347 | TGGAAAATGTGCACAACTAGGA |
| Vu6-95f | 20 702 486 | TTGTGTATTGGGTAAGTGCCAA | Vu6-95r | 20 702 508 | TCTTCCTCAGTCCCAATACTCA |
| Vu6-97f | 23 626 647 | ATTTTCTTCTTGGCTGCTGTTC | Vu6-97r | 23 626 669 | CTCTCTCCCACTCCTTTCAATG |
| Vu6-99f | 28 111 300 | CACTCCCAAGGAAGATGAGATC | Vu6-99r | 28 111 322 | GTTCTCAGGGGAAAAGGTGATT |
| Vu6-100f | 30 559 787 | ACATCTATCGCCAATCTCACAG | Vu6-100r | 30 559 809 | CAAATGAAGTGGGGAGGAAGAA |
| Vu6-101f | 31 914 466 | CACTTGAAGACGCTAAGAACAC | Vu6-101r | 31 914 488 | ACCTCCAACAAGTTCATGATCA |
| Vu6-102f | 32 539 051 | TATTGGAGAAAGGGCATAGCAG | Vu6-102r | 32 539 073 | CTTTTCACCGGTATCCCTTCTT |
| Vu7-105f | 13 121 356 | TCCTCTACCTTTCACCAAGCTA | Vu7-105r | 13 121 378 | ATGGTATGTCTTGTGATGGCAA |
| Vu7-106f | 23 354 328 | GATAGCTCACAACACAGACACT | Vu7-106r | 23 354 350 | TGTATCGGTACTCACATGCTTC |
| Vu7-108f | 29 923 695 | CACTTAGCATCTGATGGCCTTA | Vu7-108r | 29 923 717 | ACTGGGGTATTTCAAAGTGAGG |
| Vu7-109f | 31 969 597 | AAAGGGGTCACTCATACACATG | Vu7-109r | 31 969 619 | GACACCACTTTAACCCAAAACC |
| Vu7-110f | 35 390 277 | TGTGGTAGCTCAGTGTTAGTTG | Vu7-110r | 35 390 299 | AATATAACATAGGCCGGCTGAC |
| Vu8-113f | 6 256 060 | TGCGTGTAAGTGTCAGAGAAAT | Vu8-113r | 6 256 082 | AAGTGTGGTGAGGAAAGGTATG |
| Vu8-115f | 10 801 610 | TTGCTTAACCTGAGATGAACGT | Vu8-115r | 10 801 632 | ACGGGAGGTTATAGGCTATTTG |
| Vu8-116f | 21 415 000 | GACACAAGATCGAATCCAAGGA | Vu8-116r | 21 415 022 | GCACTGCATCTTTCACTTTCAT |
| Vu9-122f | 1 063 922 | CCTGGTACTTGGACTCAGATTT | Vu9-122r | 1 063 944 | CAGCTGTTGATGTTGAACCAAA |
| Vu9-123f | 2 491 158 | TTGTGTTAAAGGTGGTGCATTG | Vu9-123r | 2 491 180 | AGGGGTAAAGCAAGCAACATAT |
| Vu9-124f | 4 327 558 | GTCTCTATGGCGGTCAAGTTTA | Vu9-124r | 4 327 580 | CTTCATGATCTATCTGCCCCAG |
| Vu9-125f | 5 918 421 | GTATCGGATGCTGTTATTTGGG | Vu9-125r | 5 918 443 | GTGGAATTTCTATTTTGCCTGC |
| Vu9-126f | 7 169 197 | GATCATTGAACTCCCTTGGTCA | Vu9-126r | 7 169 219 | GTTAGCAACAGCAACCGATAAG |
| Vu9-127f | 9 039 864 | GCAGTGACCTCTCAATTCATCT | Vu9-127r | 9 039 886 | CCCTGCATCTCCATTTTGATTT |
| Vu9-129f | 15 935 910 | ACGATTCTACTGCTTAGCCATC | Vu9-129r | 15 935 932 | GCGAGAATTAAAGGAAACGTCA |
| Vu9-131f | 22 071 671 | CCCCGAATAAGTGTTCTAGGTG | Vu9-131r | 22 071 693 | CAAAACATGGCCATACATCTCG |
| Vu9-135f | 33 519 683 | CACTGCAACAATTATACGCCTT | Vu9-135r | 33 519 705 | TGTCACAGCTCCAAAGAATGAT |
| Vu9-136f | 34 865 479 | AAAGTTGTAGGCTAAGGTGGAC | Vu9-136r | 34 865 501 | CACAGATAACGAGGAGATAGGC |
| Vu9-138f | 40 694 878 | AAACCCACTCATGTACAAGACT | Vu9-138r | 40 694 900 | TGGTGTTCATCAAGATTTCACG |
| Vu10-141f | 5 081 721 | GCGATTGATTTAAGGCACAGTA | Vu10-141r | 5 081 743 | AAGTCAGGGGTTTTGATCAAGT |
| Vu10-142f | 7 985 569 | AGTCACAGTTTCAGTCACCAAA | Vu10-142r | 7 985 591 | ACGGAGATCAGTCACTATAACC |
| Vu10-144f | 12 679 952 | AGGGCTTCAATTTTCTCCTTCA | Vu10-144r | 12 679 974 | GAATTTGGAAGGCTGTTGTTGT |
| Vu10-145f | 15 994 371 | CCGGAAAACTCACAACATCATT | Vu10-145r | 15 994 393 | CCAACTTTTGAATGCTTGACCA |
| Vu10-146f | 17 013 915 | ACTGGTCTTCATTTTGTGCTGA | Vu10-146r | 17 013 937 | AGGCTTCATTGGGTAGTGTTAC |
| Vu10-147f | 23 281 009 | ACTTCATCAATGTCCACCTCTT | Vu10-147r | 23 281 031 | CGCAACGGATAGGAAATGGATA |
| Vu10-149f | 28 197 383 | TGTTTCTTGCGCTGACATAGTA | Vu10-149r | 28 197 405 | TTAGCAGTTGTCGAGTCTCTTC |
| Vu10-152f | 36 129 524 | AGTAAGCACAGTCTTAGAGCAA | Vu10-152r | 36 129 546 | TAGGCAATTCACAGTCTCTTGG |
| Vu10-154f | 39 650 768 | GACAGAAGAGGGGTTCAGAATC | Vu10-154r | 39 650 790 | AGTTCAAGTACATGCTCCAACA |
| Vu11-157f | 10 620 552 | CATTTCGCTCCCTGAAGTTTC | Vu11-157r | 10 620 573 | TGGTTTAGAAAGAGGTTACGTG |
| Vu11-160f | 24 401 745 | CCAACACGCTTTCATGATTCAA | Vu11-160r | 24 401 767 | GGTCTCGTGCAAAAGTAATGTC |
| Vu11-161f | 28 839 447 | AGTATCCAACAACCAACCATCA | Vu11-161r | 28 839 469 | GCGTGGAAATATGTATCGATCG |
| Vu11-162f | 32 266 203 | GGTCACGTCCAGAGATAATTACT | Vu11-162r | 32 266 226 | CACTTGCCAATATTCACAGCA |
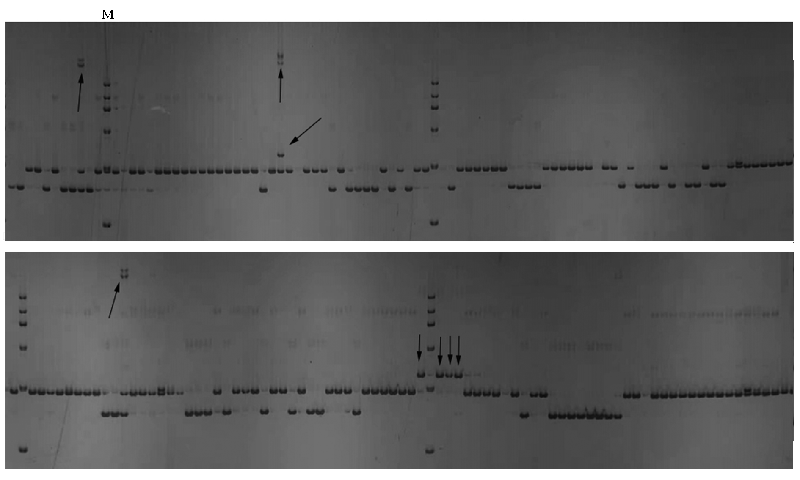
图3 引物Vu9-125在173份长豇豆材料中的扩增图 M:100 ~ 600 bp的DNA marker,“↑”代表长度 > 600 bp特异性条带,“↓”代表长度100 ~ 600 bp特异性条带。
Fig. 3 Amplification products of marker Vu6-94 in the natural population M:100-600 bp DNA markers,“↑”denotes the length of specific band longer than 600 bp,“↓”denotes the length of specific band between 100-600 bp.
| [1] | An Ji-ni. 2011. The inheritance of pod pigmentation pattern/seed coat colour and genetic diversity of cowpea(Vigna unguiculata Walp)[M. D. Dissertation]. Wuhan: Huazhong Agricultural University. (in Chinese) |
| 安吉尼. 2011. 豇豆遗传多样性及豆荚着色模式和种皮颜色的遗传分析[硕士论文]. 武汉: 华中农业大学. | |
| [2] |
Botstein D, White R L, Skolnick M, Davis R W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32 (3):314-331.
pmid: 6247908 |
| [3] | Carvalho M, Munoz-Amatriain M, Castro I, Lino-Neto T, Matos M, Egea-Cortines M, Rosa E, Close T, Carnide V. 2017. Genetic diversity and structure of Iberian Peninsula cowpeas compared to world-wide cowpeas accessions using high density SNP markers. BioMed Central Genomics, 18 (1):891. |
| [4] | Chen Chan-you, Pan Lei, Hu Zhi-hui, Peng Hai, Ding Yi. 2008. RAPD analysis of yardlong bean (Vigna unguiculata ssp. sesquipedalis)cultivars. Journal of Jianghan University(Natural Science), 36 (4):77-83. (in Chinese) |
| 陈禅友, 潘磊, 胡志辉, 彭海, 丁毅. 2008. 长豇豆品种资源的RAPD分析. 江汉大学学报(自然科学版), 36 (4):77-83. | |
| [5] |
Chen Ming-li, Wang Lan-fen, Wu Jing, Zhang Xiao-yan, Yang Guang-dong, Wang Shu-min. 2014. Development of genomic SSR markers in common bean and their transferability in asparagus bean and adzuki bean. Acta Agronomica Sinica, 40 (5):924-933. (in Chinese)
doi: 10.3724/SP.J.1006.2014.00924 URL |
| 陈明丽, 王兰芬, 武晶, 张晓艳, 杨广东, 王述民. 2014. 普通菜豆基因组SSR标记开发及在豇豆和小豆中的通用性分析. 作物学报, 40 (5):924-933. | |
| [6] |
Chen R, Chang L C, Cai X, Wu J, Liang J L, Lin R M, Song Y, Wang X W. 2021. Development of InDel markers for Brassica rapa based on a high-resolution melting curve. Horticultural Plant Journal, 7 (1):31-37.
doi: 10.1016/j.hpj.2020.05.003 URL |
| [7] | Egbadzor K F, Ofori K, Yeboah M, Aboagye L M, Opoku-Agyeman M O, Danquah E Y, Offei S K. 2014. Diversity in 113 asparagus bean [Vigna unguiculata(L)Walp] accessions assessed with 458 SNP markers. Springer Plus, 3:541. |
| [8] |
Fatokun C, Girma G, Abberton M, Gedil M, Unachukwu N, Oyatomi O, Yusuf M, Rabbi I, Boukar O. 2018. Genetic diversity and population structure of a mini-core subset from the world asparagus bean [Vigna unguiculata(L)Walp.]germplasm collection. Scientific Reports, 8 (1):16035.
doi: 10.1038/s41598-018-34555-9 URL |
| [9] | Gonzalez A M, Yuste-Lisbona F J, Rodino A P, de Ron A M, Capel C, Garcia-Alcazar M, Lozano R, Santalla M. 2015. Uncovering the genetic architecture of Colletotrichum lindemuthianum resistance through QTL mapping and epistatic interaction analysis in common bean. Frontiers in Plant Science, 6:141. |
| [10] | Guo Guang-jun, Sun Qian, Liu Jin-bing, Pan Bao-gui, Diao Wei-ping, Ge Wei, Gao Chang-zhou, Wang Shu-bin. 2015. Development and application of pepper InDel markers based on genome re-sequencing. Journal of Southern Agriculture, 31 (6):1400-1406. (in Chinese) |
| 郭广君, 孙茜, 刘金兵, 潘宝贵, 刁卫平, 戈伟, 高长洲, 王述彬. 2015. 基于辣椒基因组重测序的InDel标记开发及应用. 江苏农业学报, 31 (6):1400-1406. | |
| [11] | Hu Tao-zhu, Lin Li, Hu Ji-jun, Tai Lian-shai, Wang Sheng-jie, Liu Yang. 2019. Construction of fingerprints of tomato varieties by InDel markers. Journal of Shanghai Jiaotong University(Agricultural Science), 37 (2):24-29. (in Chinese) |
| 胡陶铸, 林丽, 胡继军, 邰连赛, 王圣洁, 刘杨. 2019. 利用InDel标记构建西红柿新品种指纹图谱. 上海交通大学学报(农业科学版), 37 (2):24-29. | |
| [12] | Ji Kang-na, Zhi Jun-jie, Lin Dan-ni, Yan Shuang-shuang, Tian Shi-bing, Cao Bi-hao, Qiu Zheng-kun. 2019. Development and application of eggplant InDel markers based on whole genome re-sequencing datasets. Journal of Plant Genetic Resources, 20 (5):1278-1288. (in Chinese) |
| 吉康娜, 郅俊杰, 林丹妮, 颜爽爽, 田时炳, 曹必好, 邱正坤. 2019. 基于茄子基因组重测序的InDel标记开发及应用. 植物遗传资源学报, 20 (5):1278-1288. | |
| [13] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. 1000 genome project data processing subgroup. The sequence alignment/map format and SAM tools. Bioinformatics, 25 (16):2078-2079.
doi: 10.1093/bioinformatics/btp352 URL |
| [14] | Li Si-geng, Shen Di, Liu Bo, Qiu Yang, Zhang Xiao-hui, Zhang Zhong-hua, Wang Hai-ping, Li Xi-xiang. 2013. Development and application of cucumber InDel markers based on genome re-sequencing. Journal of Plant Genetic Resources, 14 (2):278-283. (in Chinese) |
| 李斯更, 沈镝, 刘博, 邱杨, 张晓辉, 张忠华, 王海平, 李锡香. 2013. 基于黄瓜基因组重测序的InDel标记开发及其应用. 植物遗传资源学报, 14 (2):278-283. | |
| [15] | Li Yang. 2017. Drought tolerance evaluation and SNP molecular maekers of cucumber germplasm resources[M. D. Dissertation]. Harbin: Northeast Agricultural University. (in Chinese) |
| 李阳. 2017. 黄瓜种质资源耐旱性评价及SNP分子标记的筛选[硕士论文]. 哈尔滨: 东北农业大学. | |
| [16] |
Lonardi S, Munoz-Amatriain M, Liang Q, Shu S, Wanamaker S I, Lo S, Tanskanen J, Schulman A H, Zhu T, Luo M C, Alhakami H, Ounit R, Hasan A M, Verdier J, Roberts P A, Santos J, Ndeve A, Dolezel J, Vrana J, Hokin S A, Farmer A D, Cannon S B, Close T J. 2019. The genome of cowpea(Vigna unguiculata [L.] Walp.). Plant J, 98 (5):767-782.
doi: 10.1111/tpj.14349 URL |
| [17] |
Nagy S, Poczai P, Cernák I, Gorji AM, Hegedűs G, Taller J. 2012. PICcalc:an online program to calculate polymorphic information content for molecular genetic studies. Biochemical Genetics, 50 (9):670-672.
doi: 10.1007/s10528-012-9509-1 URL |
| [18] | Pan Lei, Li Yi, Yu Xiao-lu, Guo Rui, Chen Chan-you. 2015. Association analysis between yield trait in asparagus bean and ssr molecular markers. Hubei Agricultural Sciences, 54 (16):3952-3958. (in Chinese) |
| 潘磊, 李依, 余晓露, 郭瑞, 陈禅友. 2015. 豇豆产量性状与SSR分子标记的关联分析. 湖北农业科学, 54 (16):3952-3958. | |
| [19] |
Ravelombola W, Shi A, Weng Y, Mou B, Motes D, Clark J, Chen P, Srivastava V, Qin J, Dong L, Yang W, Bhattarai G, Sugihara Y. 2018. Association analysis of salt tolerance in asparagus bean[Vigna unguiculata(L.)Walp]at germination and seedling stages. Theoretical and Applied Genetics, 131 (1):79-91.
doi: 10.1007/s00122-017-2987-0 pmid: 28948303 |
| [20] | Tan Hua-qiang. 2014. Using morphology and RAPD, ISSR molecular markers to analyze the genetic relationship of 68 cowpea varieties[M. D. Dissertation]. Chengdu: Sichuan Agricultural University. (in Chinese) |
| 谭华强. 2014. 利用形态学和RAPD,ISSR分子标记分析68个豇豆品种的亲缘关系[硕士论文]. 成都: 四川农业大学. | |
| [21] |
Wu X, Li G, Wang B, Hu Y, Wu X, Wang Y, Lu Z, Xu P. 2018. Fine mapping Ruv2,a new rust resistance gene in asparagus bean(Vigna unguiculata),to a 193-kb region enriched with NBS-type genes. Theoretical and Applied Genetics, 131 (12):2709-2718.
doi: 10.1007/s00122-018-3185-4 URL |
| [22] | Xia Q, Pan L, Zhang R, Ni X, Wang Y, Dong X, Gao Y, Zhang Z, Kui L, Li Y, Wang W, Yang H, Chen C, Miao J, Chen W, Dong Y. 2019. The genome assembly of asparagus bean,Vigna unguiculata ssp. sesquipedialis. Scientific Data, 6 (1):124. |
| [23] | Xu P, Wu X, Wang B, Hu T, Lu Z, Liu Y, Qin D, Wang S, Li G. 2013. QTL mapping and epistatic interaction analysis in asparagus bean for several characterized and novel horticulturally important traits. BioMed Central Genetic, 14:4. |
| [24] | Xu Yan-hong, Guan Jian-ping, Zong Xu-xiao. 2007. Genetic diversity analysis of asparagus bean germplasm resources with SSR markers. Acta Agronomica Sinica, 33 (7):1206-1209. (in Chinese) |
| 徐雁鸿, 关建平, 宗绪晓. 2007. 豇豆种质资源SSR标记遗传多样性分析. 作物学报, 33 (7):1206-1209. | |
| [25] | Yue Xiao-peng. 2014. Development of InDel markers based on whole genome resequencing in Brassica napus[M. D. Dissertation]. Wuhan: Huazhong Agricultural University. (in Chinese) |
| 岳晓鹏. 2014. 基于甘蓝型油菜基因组重测序开发InDel标记[硕士论文]. 武汉: 华中农业大学. | |
| [26] | Zhang Guo-ru, Tang Ya-ping, Yang Tao, Pa Ti-gu-li, Wang Bai-ke, Li Ning, Wang Juan, Li Xiao-qin, Yu Qing-hui, Yang Sheng-bao. 2019. Development of InDel markers for tomato based on re-sequencing data. Molecular Plant Breeding, 17 (14):4692-4697. (in Chinese) |
| 张国儒, 唐亚萍, 杨涛, 帕提古丽, 王柏柯, 李宁, 王,m娟, 李晓琴, 余庆辉, 杨生保. 2019. 基于重测序西红柿InDel标记的开发. 分子植物育种, 17 (14):4692-4697. | |
| [27] | Zhang Hong-mei, Zhai Wen, Jin Hai-jun, Ding Xiao-tao, Yu Ji-zhu. 2019. Genetic diversity analysis of 23 cucumber germplasms and screening of core germplasm resources using InDel marker. Acta Agriculturae Shanghai, 35 (4):28-33. (in Chinese) |
| 张红梅, 翟文, 金海军, 丁小涛, 余纪柱. 2019. 利用InDel标记分析23份黄瓜种质的遗传多样性及核心种质资源筛选. 上海农业学报, 35 (4):28-33. |
| [1] | 王 瑞, 洪文娟, 罗 华, 赵丽娜, 陈 颖, 王 君, . 石榴品种SSR指纹图谱构建及杂种父本鉴定[J]. 园艺学报, 2023, 50(2): 265-278. |
| [2] | 王梦梦, 孙德岭, 陈 锐, 杨迎霞, 张 冠, 吕明杰, 王 倩, 谢添羽, 牛国保, 单晓政, 谭 津, 姚星伟, . 花椰菜核心种质的构建与评价[J]. 园艺学报, 2023, 50(2): 421-431. |
| [3] | 汤雨晴, 杨惠栋, 闫承璞, 王斯妤, 王雨亭, 胡钟东, 朱方红. 基于重测序的‘金兰柚’基因组InDel标记的开发及应用[J]. 园艺学报, 2023, 50(1): 15-26. |
| [4] | 吕正鑫, 贺艳群, 贾东峰, 黄春辉, 钟敏, 廖光联, 朱壹, 袁开昌, 刘传浩, 徐小彪. 猕猴桃种质资源表型性状遗传多样性分析[J]. 园艺学报, 2022, 49(7): 1571-1581. |
| [5] | 李文婷, 李翠晓, 林小清, 郑永钦, 郑正, 邓晓玲. 基于STR位点对广东省柑橘溃疡病菌种群遗传结构的分析[J]. 园艺学报, 2022, 49(6): 1233-1246. |
| [6] | 张萌, 单玉莹, 杨业波, 翟飞飞, 王兆山, 巨关升, 孙振元, 李振坚. 中国石斛属植物遗传资源的AFLP分析[J]. 园艺学报, 2022, 49(6): 1339-1350. |
| [7] | 聂兴华, 李伊然, 田寿乐, 王雪峰, 苏淑钗, 曹庆芹, 邢宇, 秦岭. 中国板栗品种(系)DNA指纹图谱构建及其遗传多样性分析[J]. 园艺学报, 2022, 49(11): 2313-2324. |
| [8] | 李世帆, 邵宇纯, 祁豪男, 杨兰, 李小伟, 徐秉良, 陈雅寒. 甘肃省苹果坏死花叶病毒检测及遗传多样性分析[J]. 园艺学报, 2022, 49(11): 2431-2438. |
| [9] | 陈明堃, 陈璐, 孙维红, 马山虎, 兰思仁, 彭东辉, 刘仲健, 艾叶. 建兰种质资源遗传多样性分析及核心种质构建[J]. 园艺学报, 2022, 49(1): 175-186. |
| [10] | 李艳红, 聂俊, 郑锦荣, 谭德龙, 张长远, 史亮亮, 谢玉明. 华南地区樱桃番茄表型性状遗传多样性分析及综合评价[J]. 园艺学报, 2021, 48(9): 1717-1730. |
| [11] | 赵玉靖, 卢银, 冯大领, 罗蕾, 吴芳, 杨锐, 陈悦琦, 孙晓雪, 王彦华, 陈雪平, 申书兴, 罗双霞, 赵建军. 大白菜自交系背景选择InDel标记与重要农艺性状的关联分析[J]. 园艺学报, 2021, 48(7): 1282-1294. |
| [12] | 赵青, 都真真, 李锡香, 宋江萍, 张晓辉, 阳文龙, 贾会霞, 王海平. 利用SSRseq分子标记的大蒜种质资源遗传多样性研究[J]. 园艺学报, 2021, 48(7): 1397-1408. |
| [13] | 江锡兵, 章平生, 徐阳, 吴聪连, 张东北, 龚榜初, 吴开云, 赖俊声. 栗杂交F1代SSR标记遗传多样性分析[J]. 园艺学报, 2021, 48(5): 897-907. |
| [14] | 王振江, 罗国庆, 戴凡炜, 肖更生, 林森, 李智毅, 唐翠明. 基于8个农艺性状的569份果桑种质遗传多样性分析[J]. 园艺学报, 2021, 48(12): 2375-2384. |
| [15] | 高 源,王大江,王 昆*,丛佩华*,张彩霞,李连文,朴继成. 基于叶绿体DNA分析的楸子种质遗传多样性研究[J]. 园艺学报, 2020, 47(5): 853-863. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司