
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (3): 545-559.doi: 10.16420/j.issn.0513-353x.2023-0098
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
LIU Jinhong, WANG Zheng, YU Hao, XIN Yirui, QI Guoning, LIU Shenkui, REN Huimin*( )
)
Received:2023-04-16
Revised:2023-11-17
Online:2024-03-25
Published:2024-03-22
Contact:
REN Huimin
LIU Jinhong, WANG Zheng, YU Hao, XIN Yirui, QI Guoning, LIU Shenkui, REN Huimin. Identification of SLAC Gene Family in Phyllostachys edulis and Characterization of PheSLAC1[J]. Acta Horticulturae Sinica, 2024, 51(3): 545-559.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0098
| 引物用途 Primer use | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | |
|---|---|---|---|
| 实时荧光定量引物 qRT-PCR primer | NTB | F:TCTTGTTTGACACCGAAGAGGAG; | R:AATAGCTGTCCCTGGAGGAGTTT |
| PheSLAC1.1 | F:FTTCTTCATGGCGCTCTTCTT; | R:GACATGAGGGAGAGGCTCAG | |
| PheSLAC1.2 | F:GCGCGGACATTATTCTTCAT; | R:GACATGAGGGAGAGGCTCAG | |
| PH02Gene35134 | F:ATCTACGGCCAGTGGTTCAC; | R:TGACGAACAGCACGAGGTAG | |
| PH02Gene08796 | F:CATGAACTACCTCTTCGCGC; | R:GAACCACTGGCCGTAGATCT | |
| PH02Gene45054 | F:ACTACAGCGTGCTGTTCGTG; | R:GAACCCACGGAAGAAGTTGA | |
| PH02Gene22907 | F:TGTTCGTGACGCTCTACCAG; | R:TGAACCCCCTGAAAAAGTTG | |
| PH02Gene22131 | F:CCAGAGTCCAACGCTTCTTC; | R:ATCTTGAGCGTGATGTGCAG | |
| PH02Gene10869 | F:AGCTAAGGTCGGAGGAGGAG; | R:ATCTTGAGCGTGATGTGCAG | |
| PH02Gene20948 | F:ATCTACGGGCAGTGGATGTC; | R:TGGTACAGCGTGACGAAGAG | |
| PH02Gene48421 | F:GCCTCCCTGAATGAGCATAA; | R:ATGCTGAGCGAAAGGGAGTA | |
| PH02Gene25481 | F:GTGGATGAACAGGGGAAAGA; | R:GGACCTCGAGTTTTGTCTCG | |
| PH02Gene41494 | F:GGAAGGACGTCGTGATTAGC; | R:GGACCTCGAGTTTTGTCTCG | |
| PH02Gene06334 | F:CTGTCCGCCGAGAGTTTTAC; | R:GAGCTCAAGGCAGAAGATGG | |
| PH02Gene06335 | F:GGCTTCGTCTCGGTGATCTA; | R:CATCCACTGCCCGTAGATCT | |
| PH02Gene20553 | F:ATCTACGGGCAGTGGATGTC; | R:TGACGAACAGCACGAGGTAG | |
| AtACTIN2 | F:GGTAACATTGTGCTCAGTGGTGG; | R:AACGACCTTAATCTTCATGCTGC | |
| 转基因和亚细胞定位 Transgene and subcellular localization | PheSLAC1.1-1304 | F: GAAGATCTTATGGCAGCCGAGCCATCGTC R: CGGACTAGTGTCTGTTTTCTCCTCTTCGTCCTT | |
| PheSLAC1.2-1304 | F: GAAGATCTTATGGCAGCCGAGCCATCGTC R: CGGACTAGTTTCTGTTTCCTCCTCTTCATCCTTG | ||
| PheSLAC1.1-GFP | F: TCCCCCCGGGATGGCAGCCGAGCCATCGTCT R: CTAGTCTAGAGTCTGTTTTCTCCTCTTCGTCCTT | ||
| PheSLAC1.2-GFP | F: TCCCCCCGGGATGGCAGCCGAGCCATCGTC R: CTAGTCT AGATTCTGTTTCCTCCTCTTCATCCT | ||
Table 1 Primers used in this study
| 引物用途 Primer use | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence | |
|---|---|---|---|
| 实时荧光定量引物 qRT-PCR primer | NTB | F:TCTTGTTTGACACCGAAGAGGAG; | R:AATAGCTGTCCCTGGAGGAGTTT |
| PheSLAC1.1 | F:FTTCTTCATGGCGCTCTTCTT; | R:GACATGAGGGAGAGGCTCAG | |
| PheSLAC1.2 | F:GCGCGGACATTATTCTTCAT; | R:GACATGAGGGAGAGGCTCAG | |
| PH02Gene35134 | F:ATCTACGGCCAGTGGTTCAC; | R:TGACGAACAGCACGAGGTAG | |
| PH02Gene08796 | F:CATGAACTACCTCTTCGCGC; | R:GAACCACTGGCCGTAGATCT | |
| PH02Gene45054 | F:ACTACAGCGTGCTGTTCGTG; | R:GAACCCACGGAAGAAGTTGA | |
| PH02Gene22907 | F:TGTTCGTGACGCTCTACCAG; | R:TGAACCCCCTGAAAAAGTTG | |
| PH02Gene22131 | F:CCAGAGTCCAACGCTTCTTC; | R:ATCTTGAGCGTGATGTGCAG | |
| PH02Gene10869 | F:AGCTAAGGTCGGAGGAGGAG; | R:ATCTTGAGCGTGATGTGCAG | |
| PH02Gene20948 | F:ATCTACGGGCAGTGGATGTC; | R:TGGTACAGCGTGACGAAGAG | |
| PH02Gene48421 | F:GCCTCCCTGAATGAGCATAA; | R:ATGCTGAGCGAAAGGGAGTA | |
| PH02Gene25481 | F:GTGGATGAACAGGGGAAAGA; | R:GGACCTCGAGTTTTGTCTCG | |
| PH02Gene41494 | F:GGAAGGACGTCGTGATTAGC; | R:GGACCTCGAGTTTTGTCTCG | |
| PH02Gene06334 | F:CTGTCCGCCGAGAGTTTTAC; | R:GAGCTCAAGGCAGAAGATGG | |
| PH02Gene06335 | F:GGCTTCGTCTCGGTGATCTA; | R:CATCCACTGCCCGTAGATCT | |
| PH02Gene20553 | F:ATCTACGGGCAGTGGATGTC; | R:TGACGAACAGCACGAGGTAG | |
| AtACTIN2 | F:GGTAACATTGTGCTCAGTGGTGG; | R:AACGACCTTAATCTTCATGCTGC | |
| 转基因和亚细胞定位 Transgene and subcellular localization | PheSLAC1.1-1304 | F: GAAGATCTTATGGCAGCCGAGCCATCGTC R: CGGACTAGTGTCTGTTTTCTCCTCTTCGTCCTT | |
| PheSLAC1.2-1304 | F: GAAGATCTTATGGCAGCCGAGCCATCGTC R: CGGACTAGTTTCTGTTTCCTCCTCTTCATCCTTG | ||
| PheSLAC1.1-GFP | F: TCCCCCCGGGATGGCAGCCGAGCCATCGTCT R: CTAGTCTAGAGTCTGTTTTCTCCTCTTCGTCCTT | ||
| PheSLAC1.2-GFP | F: TCCCCCCGGGATGGCAGCCGAGCCATCGTC R: CTAGTCT AGATTCTGTTTCCTCCTCTTCATCCT | ||
| 种类 Species | 蒸腾速率/(mmol · m-2 · s-1) Transpiration rate | 气孔导度/(mmol · m-2 · s-1) Stomatal conductance |
|---|---|---|
| 毛竹 Phyllostachys edulis | 0.938 | 0.058 |
| 菲黄竹 Pleioblastus viridistriatus | 1.198 | 0.073 |
| 绿槽罗汉竹 Phyllostachys aureosulcata | 0.592 | 0.037 |
| 唐竹 Sinobambusa tootsik | 0.482 | 0.029 |
| 紫竹 Phyllostachys nigra | 0.641 | 0.040 |
| 橄榄竹 Indosasa gigantea | 0.655 | 0.048 |
| 凤尾竹 Bambusa multiplex | 1.562 | 0.101 |
Table 2 Transpiration rate and stomatal conductance of different bamboo species
| 种类 Species | 蒸腾速率/(mmol · m-2 · s-1) Transpiration rate | 气孔导度/(mmol · m-2 · s-1) Stomatal conductance |
|---|---|---|
| 毛竹 Phyllostachys edulis | 0.938 | 0.058 |
| 菲黄竹 Pleioblastus viridistriatus | 1.198 | 0.073 |
| 绿槽罗汉竹 Phyllostachys aureosulcata | 0.592 | 0.037 |
| 唐竹 Sinobambusa tootsik | 0.482 | 0.029 |
| 紫竹 Phyllostachys nigra | 0.641 | 0.040 |
| 橄榄竹 Indosasa gigantea | 0.655 | 0.048 |
| 凤尾竹 Bambusa multiplex | 1.562 | 0.101 |
| 基因登录号 ID | 染色体位置 Chromosome location | 等电点 pI | 相对分子量/kD MW | 长度/aa Length | 跨膜区数量 No. of transmembrane domain |
|---|---|---|---|---|---|
| PH02Gene00592.t1 | hic_scaffold_24 | 9.63 | 61 635.52 | 558 | 9 |
| PH02Gene01933.t1 | hic_scaffold_23 | 9.94 | 62 657.88 | 571 | 10 |
| PH02Gene35134.t1 | hic_scaffold_16 | 10.18 | 33 127.26 | 294 | 9 |
| PH02Gene08796.t1 | hic_scaffold_6 | 9.87 | 42 370.12 | 384 | 10 |
| PH02Gene45054.t2 | hic_scaffold_9 | 9.13 | 64 409.35 | 586 | 10 |
| PH02Gene22907.t1 | hic_scaffold_18 | 8.52 | 67 869.30 | 621 | 10 |
| PH02Gene22131.t1 | hic_scaffold_16 | 9.26 | 64 301.36 | 583 | 10 |
| PH02Gene10869.t1 | hic_scaffold_14 | 9.18 | 67 749.68 | 609 | 10 |
| PH02Gene20948.t2 | hic_scaffold_16 | 8.82 | 72 183.69 | 651 | 10 |
| PH02Gene48421.t1 | hic_scaffold_14 | 8.70 | 68 667.45 | 613 | 10 |
| PH02Gene25481.t1 | hic_scaffold_342 | 8.02 | 73 583.66 | 663 | 9 |
| PH02Gene41494.t1 | hic_scaffold_9 | 7.71 | 69 005.90 | 623 | 9 |
| PH02Gene06334.t1 | hic_scaffold_7 | 7.03 | 70 670.36 | 640 | 8 |
| PH02Gene06335.t1 | hic_scaffold_7 | 7.30 | 70 425.26 | 640 | 10 |
| PH02Gene20553.t1 | hic_scaffold_13 | 7.09 | 77 827.50 | 708 | 9 |
Table 3 The SLAC1 gene family of moso bamboo
| 基因登录号 ID | 染色体位置 Chromosome location | 等电点 pI | 相对分子量/kD MW | 长度/aa Length | 跨膜区数量 No. of transmembrane domain |
|---|---|---|---|---|---|
| PH02Gene00592.t1 | hic_scaffold_24 | 9.63 | 61 635.52 | 558 | 9 |
| PH02Gene01933.t1 | hic_scaffold_23 | 9.94 | 62 657.88 | 571 | 10 |
| PH02Gene35134.t1 | hic_scaffold_16 | 10.18 | 33 127.26 | 294 | 9 |
| PH02Gene08796.t1 | hic_scaffold_6 | 9.87 | 42 370.12 | 384 | 10 |
| PH02Gene45054.t2 | hic_scaffold_9 | 9.13 | 64 409.35 | 586 | 10 |
| PH02Gene22907.t1 | hic_scaffold_18 | 8.52 | 67 869.30 | 621 | 10 |
| PH02Gene22131.t1 | hic_scaffold_16 | 9.26 | 64 301.36 | 583 | 10 |
| PH02Gene10869.t1 | hic_scaffold_14 | 9.18 | 67 749.68 | 609 | 10 |
| PH02Gene20948.t2 | hic_scaffold_16 | 8.82 | 72 183.69 | 651 | 10 |
| PH02Gene48421.t1 | hic_scaffold_14 | 8.70 | 68 667.45 | 613 | 10 |
| PH02Gene25481.t1 | hic_scaffold_342 | 8.02 | 73 583.66 | 663 | 9 |
| PH02Gene41494.t1 | hic_scaffold_9 | 7.71 | 69 005.90 | 623 | 9 |
| PH02Gene06334.t1 | hic_scaffold_7 | 7.03 | 70 670.36 | 640 | 8 |
| PH02Gene06335.t1 | hic_scaffold_7 | 7.30 | 70 425.26 | 640 | 10 |
| PH02Gene20553.t1 | hic_scaffold_13 | 7.09 | 77 827.50 | 708 | 9 |

Fig. 3 Phylogenetic tree(A),conserved motif(B)and conserved domains(C)in PheSLAC gene family At:Arabidopsis thaliana;Os:Oryza sativa;PH:Phyllostachys edulis. The same below.

Fig. 4 Key amino acids sites on PheSLAC1 proteins The orange label represents the phosphorylation site,the red label represents the ion selection site,and the green label represents the benzene ring.
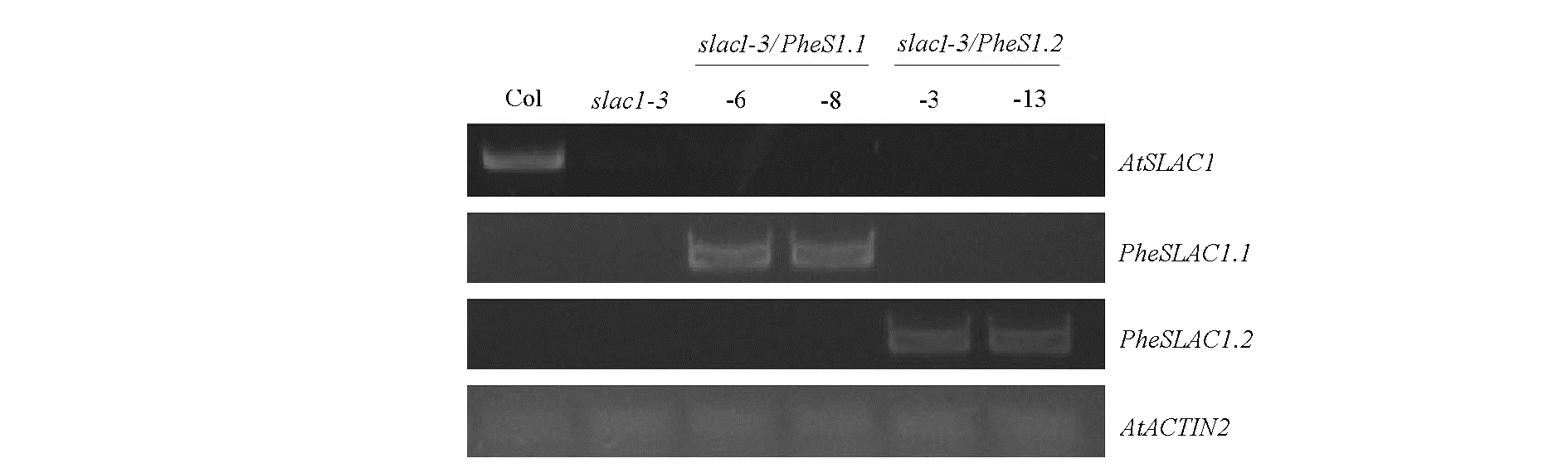
Fig. 7 Characterization of PheSLAC overexpression transgenic lines Arabidopsis wild type Col and slac1-3 were used as the control,ACTIN 2 was used as an internal standard.
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
doi: 10.1093/mp/ssm022 pmid: 19825533 |
| [38] |
|
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
|
程明圣, 邹欢欢, 黄孝风, 陈慧晶, 邹娜, 杨清培. 2021. 干旱与盐胁迫对毛竹幼苗生长及生理的影响. 江西农业大学学报, 43 (4):842-852.
|
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
doi: 10.1111/nph.13435 pmid: 25932909 |
| [12] |
doi: 10.1111/nph.14685 pmid: 28722226 |
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
doi: 10.1016/j.cub.2015.01.067 pmid: 25802151 |
| [21] |
|
| [22] |
doi: 10.1093/pcp/pcf081 pmid: 12091719 |
| [23] |
|
| [24] |
|
| [25] |
|
| [39] |
|
|
应叶青, 郭璟, 魏建芬, 姜琴, 解楠楠. 2011. 干旱胁迫对毛竹幼苗生理特性的影响. 生态学杂志, 30 (2):262-266.
|
|
| [40] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
pmid: 16101906 |
| [31] |
pmid: 12232336 |
| [32] |
|
| [33] |
|
| [1] | WU Xiangqi, SUN Li, YU Zheping, YU Qinpei, LIANG Senmiao, ZHENG Xiliang, QI Xingjiang, ZHANG Shuwen. Functional Study of MrSPL4 Gene in Response to Drought and Low Temperature Stress in Chinese Bayberry [J]. Acta Horticulturae Sinica, 2024, 51(5): 927-938. |
| [2] | ZHONG Zhaojiang, WU Zhen, ZHOU Rong, ZHU Weimin, YANG Xuedong, YU Xiaowei, XU Yan, GAO Yangyang, and JIANG Fangling. Study on the Regulatory Mechanism of SlPL Gene Affecting Tomato Fruit Cracking [J]. Acta Horticulturae Sinica, 2024, 51(2): 295-308. |
| [3] | CHEN Xin, WU Xiaolong, LIU Shengrui, HU Xianchun, and LIU Chunyan, . Effects of AMF on Photosynthetic Characteristics and Gene Expressions of Tea Plants Under Drought Stress [J]. Acta Horticulturae Sinica, 2024, 51(10): 2358-2370. |
| [4] | TIAN Jie, ZHOU Qianyi, TIE Yuanyu, SUN Haihong, and HUANG Sijie. Metabolomic Analysis of Garlic Seedling Under Drought Stress [J]. Acta Horticulturae Sinica, 2024, 51(1): 133-144. |
| [5] | WANG Yixi, YAN Shuangshuang, YU Bingwei, GAN Yuwei, QIU Zhengkun, ZHU Zhangsheng, CHEN Changming, CAO Bihao. Screening and Identification of E3 Ubiquitin Ligase Genes Relate to Bacterial Wilt Resistance in Eggplant [J]. Acta Horticulturae Sinica, 2023, 50(10): 2271-2287. |
| [6] | QIU Ziwen, LIU Linmin, LIN Yongsheng, LIN Xiaojie, LI Yongyu, WU Shaohua, YANG Chao. Cloning and Functional Analysis of the MbEGS Gene from Melaleuca bracteata [J]. Acta Horticulturae Sinica, 2022, 49(8): 1747-1760. |
| [7] | PENG Yi, LI Yuanhui, YANG Rui, ZHANG Ziyi, LI Yanan, HAN Yunhao, ZHAO Wenchao, WANG Shaohui. The Jasmonic Acid Synthesis Gene LoxD Participates in the Regulation of Tomato Drought Resistance [J]. Acta Horticulturae Sinica, 2022, 49(2): 319-331. |
| [8] | ZHANG Qian, YANG Nan, SANG Haiyu, ZHAO Rong, SONG Xiaoxi, CHEN Longqing, XIANG Lin, ZHAO Kaige. Cloning and Functional Analysis of CpTT8 Related to Anthocyanin Synthesis in Wintersweet(Chimonanthus praecox) [J]. Acta Horticulturae Sinica, 2021, 48(10): 1945-1955. |
| [9] | ZHANG Zhongxing, CHENG Li, WANG Shuangcheng, ZHANG De, LIU Bing, WANG Yanxiu. Cloning of MhMYB114-Like from Malus halliana and Its Functional Identification of Iron Deficiency Tolerance in Apple Callus [J]. Acta Horticulturae Sinica, 2021, 48(1): 127-136. |
| [10] | HUANG Yaoyao1,DENG Minghua2,PENG Chunxiu2,and WEN Jinfen1,*. Studies on the Response of Lily Petal Antioxidant Enzyme System to Drought Stress [J]. ACTA HORTICULTURAE SINICA, 2020, 47(4): 788-796. |
| [11] | SONG Yang,LIU Hongdi,WANG Haibo,ZHANG Hongjun*,and LIU Fengzhi*. Molecular Cloning and Functional Characterization of Anthocyanin Synthesis Related Genes VcTTG1 of Blueberry [J]. ACTA HORTICULTURAE SINICA, 2019, 46(7): 1270-1278. |
| [12] | YANG Guanxian,XU Haifeng,ZHANG Jing,WANG Nan,FANG Hongcheng,JIANG Shenghui,WANG Yicheng,SU Mengyu,and CHEN Xuesen*. Functional Identification of Apple Anthocyanin Regulatory Gene MdMYB111 [J]. ACTA HORTICULTURAE SINICA, 2019, 46(5): 832-840. |
| [13] | WANG Xiaodi,JI Xiaohao,ZHENG Xiaocui,WANG Yingying,SONG Yang*,and LIU Fengzhi*. Cloning and Functional Analysis of a Cold Treatment Response Factor Gene PdCIbHLH in Peach [J]. ACTA HORTICULTURAE SINICA, 2019, 46(3): 444-452. |
| [14] | PENG Liyun,WANG Yun,SUN Xueli,WANG Xiao,ZHAO Chunli,WANG Congqiao,XIANG Leilei,CHEN Jialan,LAI Zhongxiong*,and LIU Shengcai*. Expression and Functional Analysis of AmMYB2 Related to Betalain Metabolism of Amaranthus tricolor [J]. ACTA HORTICULTURAE SINICA, 2019, 46(3): 473-485. |
| [15] | WANG Chunxia,YIN Yue,WANG Zhiping,YAN Rui,Fu Linlan,and SUN Hongmei*. Effects of Regeneration and Abiotic Stress Tolerance in Polyploid Lilium davidii var. unicolor and Lilium pumilum in Vitro [J]. ACTA HORTICULTURAE SINICA, 2019, 46(12): 2359-2368. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd