
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (10): 2249-2262.doi: 10.16420/j.issn.0513-353x.2022-0585
• Research Papers • Previous Articles Next Articles
YANG Bo1, WEI Jia1, LI Kunfeng2, WANG Chengliang3, NI Junbei1, TENG Yuanwen1,4, and BAI Songling1,*( )
)
Received:2022-06-08
Revised:2022-08-22
Online:2022-10-25
Published:2022-10-31
Contact:
and BAI Songling
E-mail:songlingbai@zju.edu.cn
CLC Number:
YANG Bo, WEI Jia, LI Kunfeng, WANG Chengliang, NI Junbei, TENG Yuanwen, and BAI Songling. PpyERF060-PpyABF3-PpyMADS71 Regulates Ethylene Signaling Pathway- Mediated Pear Bud Dormancy Process[J]. Acta Horticulturae Sinica, 2022, 49(10): 2249-2262.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-0585
| 用途 Use | 引物 Primer | 序列 Sequence |
|---|---|---|
| 定量表达 | Q-PpyERF060-F | TCGGATTCTCAGTGGGACGA |
| qRT-PCR | Q-PpyERF060-R | TTACAGCATCACCACGCCTT |
| Q-PpyMADS71-F | AACAACCAGCTAAGCCAGAAG | |
| Q-PpyMADS71-R | TCAAGGGTTGGAGAAGGTGG | |
| Q-PpyABF3-F | CCGGTTCGAATTTGCCACAG | |
| Q-PpyABF3-R | CGGTTGTTGAAAGCCGGAAG | |
| 亚细胞定位 | PpyERF060-GFP-F | AGCTCGGTACCCGGGATGGCAACATCCATAGATCTTTAC |
| Subcellular localization | PpyERF060-GFP-R | CATGTCGACTCTAGAGATAGCAGACCAATCAATCTCCA |
| 酵母单杂 | pMADS71-pAbAi-F | GAAAAGCTTGAATTCGAGCTCGGGTGACTGGTTGCTGGTTGTT |
| Y1H assay | pMADS71-mutDRE1-pAbAi-R | ACGTGGAGTTTTTCACACCT |
| pMADS71-mutDRE1-pAbAi-F | AGGTGTGAAAAACTCCACGT | |
| pMADS71-mutDRE2-pAbAi-R | GTTTATTGTTTTTTTAGCCT | |
| pMADS71-mutDRE2-pAbAi-F | AGGCTAAAAAAACAATAAAC | |
| pMADS71-pAbAi-R | AGCACATGCCTCGAGGTCGACCAACGGTGATCGATGCGGTTGG | |
| 双荧光素酶 | pMADS71-all-LUC-F | TCGAGGTCGACGGTATCGATACCTAAAGCGCAACCGTAACTG |
| Dual-Luciferase assay | pMADS71-all-LUC-R | CCGCTCTAGAACTAGTGGATCCTTCTCTTTCTTCCTACCCCCT |
| PpyERF060-SK-F | GCGGCCGCTCTAGAACTAGTGATGGCAACATCCATAGATCTTTAC | |
| PpyERF060-SK-R | GGTCGACGGTATCGATAAGCTTTAGATAGCAGACCAATCAATCTC | |
| pMADS71-delDRE1-LUC-F | TCGAGGTCGACGGTATCGATATCCACGTGAACTCGGATAAACACC | |
| pMADS71-mutDRE1-LUC-R | ACGTGGAGTTTTTCACACCT | |
| pMADS71-mutDRE1-LUC-F | AGGTGTGAAAAACTCCACGT | |
| pERF060-LUC-F | TCGAGGTCGACGGTATCGATATAAAAAAGTTAGGGCTAAAATTGT | |
| pERF060-LUC-R | CCGCTCTAGAACTAGTGGATCTTAGATTTAGTCACAAAAGATAAA | |
| PpyABF3-SK-F | GCGGCCGCTCTAGAACTAGTGATGGGGTCTAATTTCAACTTCAA | |
| PpyABF3-SK-R | GGTCGACGGTATCGATAAGCTTTACCAAGGGCCTGTCAATGT | |
| PpyERF060过表达 | PpyERF060-1301-F | AGAACACGGGGGACTCTTGACATGGCAACATCCATAGATCTTTAC |
| PpyERF060 overexpression | PpyERF060-1301-R | GGGGAAATTCGAGCTGGTCACTTAGATAGCAGACCAATCAATCTC |
Table 1 Primer sequences
| 用途 Use | 引物 Primer | 序列 Sequence |
|---|---|---|
| 定量表达 | Q-PpyERF060-F | TCGGATTCTCAGTGGGACGA |
| qRT-PCR | Q-PpyERF060-R | TTACAGCATCACCACGCCTT |
| Q-PpyMADS71-F | AACAACCAGCTAAGCCAGAAG | |
| Q-PpyMADS71-R | TCAAGGGTTGGAGAAGGTGG | |
| Q-PpyABF3-F | CCGGTTCGAATTTGCCACAG | |
| Q-PpyABF3-R | CGGTTGTTGAAAGCCGGAAG | |
| 亚细胞定位 | PpyERF060-GFP-F | AGCTCGGTACCCGGGATGGCAACATCCATAGATCTTTAC |
| Subcellular localization | PpyERF060-GFP-R | CATGTCGACTCTAGAGATAGCAGACCAATCAATCTCCA |
| 酵母单杂 | pMADS71-pAbAi-F | GAAAAGCTTGAATTCGAGCTCGGGTGACTGGTTGCTGGTTGTT |
| Y1H assay | pMADS71-mutDRE1-pAbAi-R | ACGTGGAGTTTTTCACACCT |
| pMADS71-mutDRE1-pAbAi-F | AGGTGTGAAAAACTCCACGT | |
| pMADS71-mutDRE2-pAbAi-R | GTTTATTGTTTTTTTAGCCT | |
| pMADS71-mutDRE2-pAbAi-F | AGGCTAAAAAAACAATAAAC | |
| pMADS71-pAbAi-R | AGCACATGCCTCGAGGTCGACCAACGGTGATCGATGCGGTTGG | |
| 双荧光素酶 | pMADS71-all-LUC-F | TCGAGGTCGACGGTATCGATACCTAAAGCGCAACCGTAACTG |
| Dual-Luciferase assay | pMADS71-all-LUC-R | CCGCTCTAGAACTAGTGGATCCTTCTCTTTCTTCCTACCCCCT |
| PpyERF060-SK-F | GCGGCCGCTCTAGAACTAGTGATGGCAACATCCATAGATCTTTAC | |
| PpyERF060-SK-R | GGTCGACGGTATCGATAAGCTTTAGATAGCAGACCAATCAATCTC | |
| pMADS71-delDRE1-LUC-F | TCGAGGTCGACGGTATCGATATCCACGTGAACTCGGATAAACACC | |
| pMADS71-mutDRE1-LUC-R | ACGTGGAGTTTTTCACACCT | |
| pMADS71-mutDRE1-LUC-F | AGGTGTGAAAAACTCCACGT | |
| pERF060-LUC-F | TCGAGGTCGACGGTATCGATATAAAAAAGTTAGGGCTAAAATTGT | |
| pERF060-LUC-R | CCGCTCTAGAACTAGTGGATCTTAGATTTAGTCACAAAAGATAAA | |
| PpyABF3-SK-F | GCGGCCGCTCTAGAACTAGTGATGGGGTCTAATTTCAACTTCAA | |
| PpyABF3-SK-R | GGTCGACGGTATCGATAAGCTTTACCAAGGGCCTGTCAATGT | |
| PpyERF060过表达 | PpyERF060-1301-F | AGAACACGGGGGACTCTTGACATGGCAACATCCATAGATCTTTAC |
| PpyERF060 overexpression | PpyERF060-1301-R | GGGGAAATTCGAGCTGGTCACTTAGATAGCAGACCAATCAATCTC |
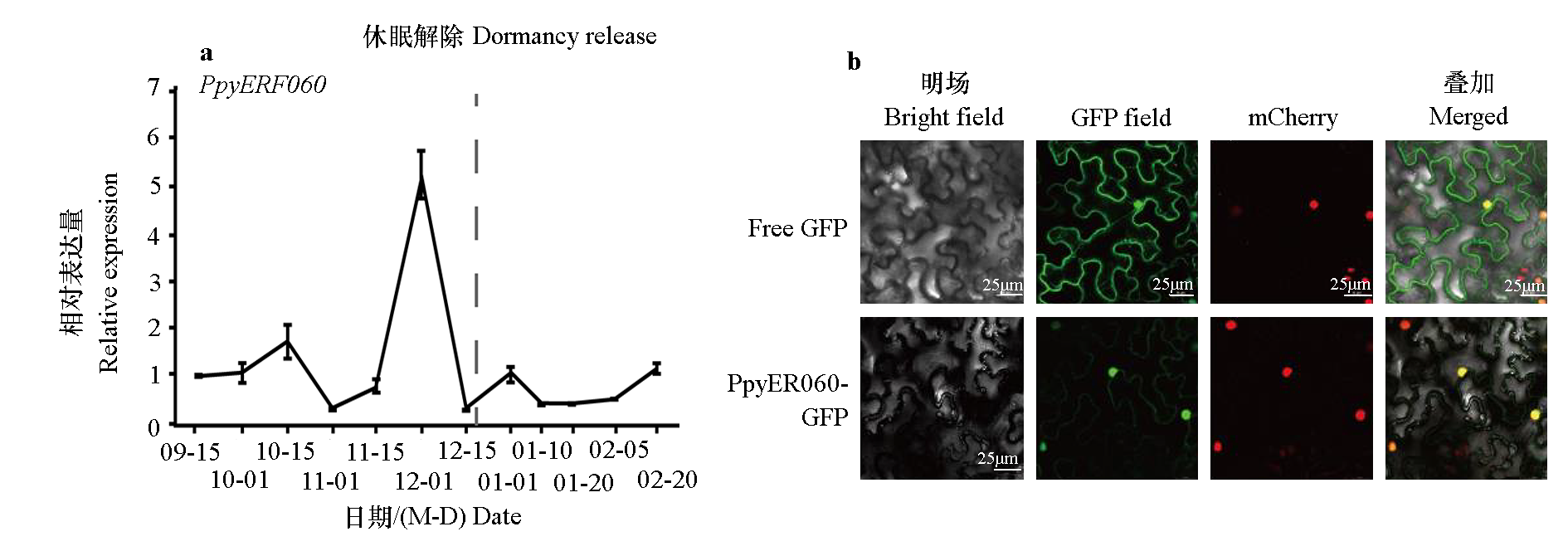
Fig. 1 The expression patterns of PpyERF060 during dormancy cycle(a)and subcellular localization of PpyERF060(b) Free GFP is used as the control. The red fluorescence indicates the location of the nucleus,and the green fluorescence indicates the location of the fusion protein. If the two fluorescence signals are in the same place,the fluorescence signal will be yellow after superposition.
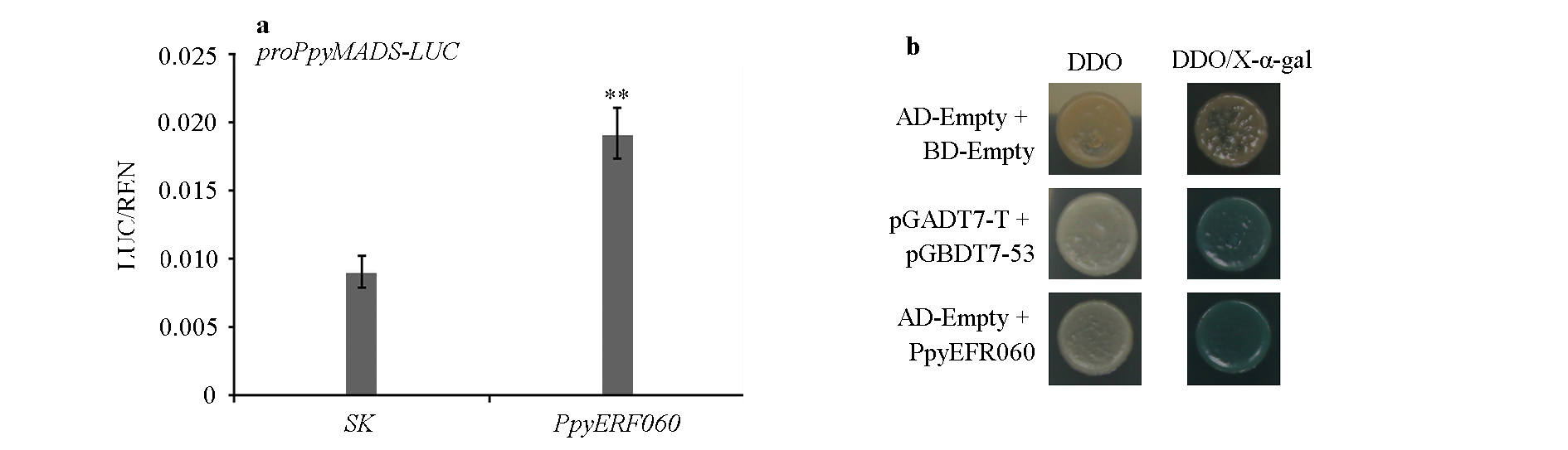
Fig. 2 The regulation pattern of PpyERF060 to PpyMADS71 expression(a)and yeast transcriptional activation assay of PpyERF060(b) Student's t-test,**P < 0.01.
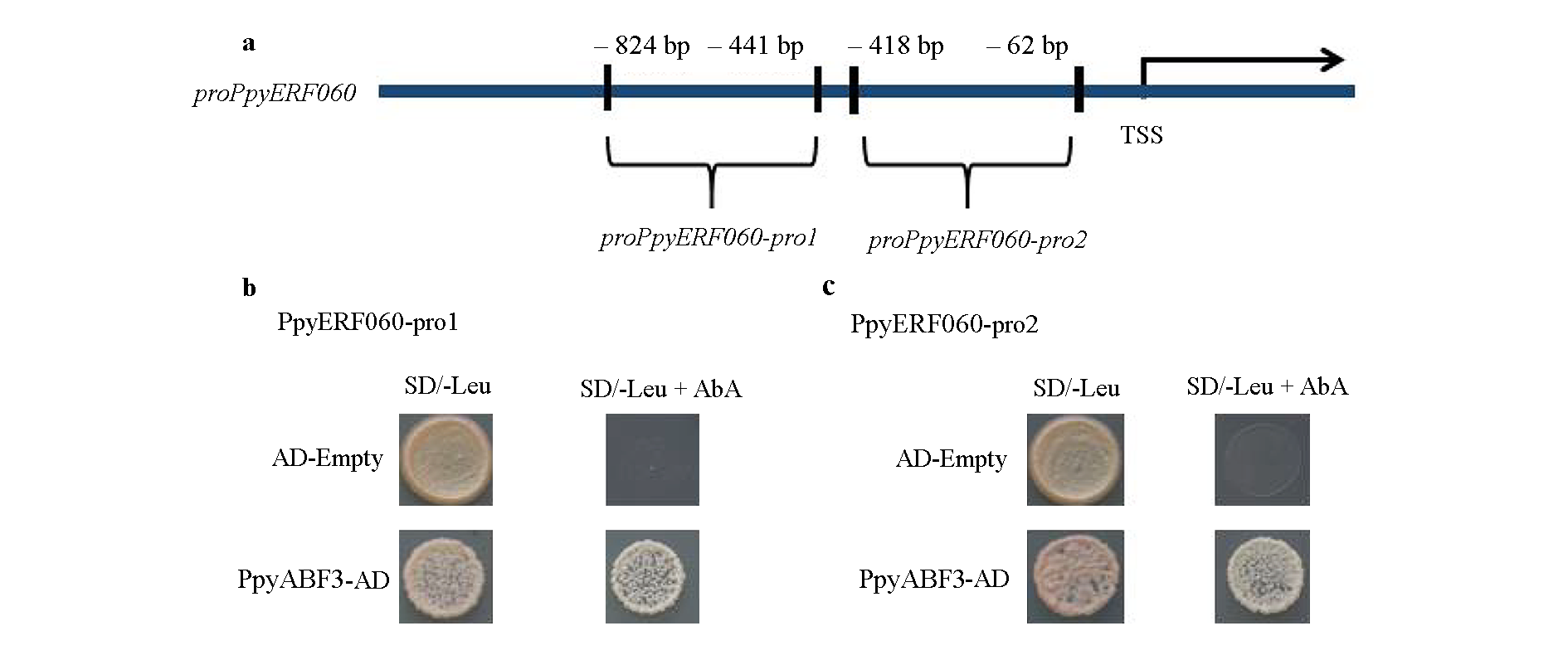
Fig. 8 The potential binding site of PpyABF3 on the promoter of PpyERF060(a)and Y1H confirmed the interaction between PpyABF3 and the promoter of PpyERF060(b)
| [1] | Arc E, Sechet J, Corbineau F, Rajjou L, Marion-Poll A. 2013. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Frontiers in Plant Science, 3 (4):63. |
| [2] |
Arora R, Rowland L J, Tanino K. 2003. Induction and release of bud dormancy in woody perennials:a science comes of age. HortScience, 38 (5):911-921.
doi: 10.21273/HORTSCI.38.5.911 URL |
| [3] |
Baldocchi D, Wong S. 2008. Accumulated winter chill is decreasing in the fruit growing regions of California. Climatic Change, 87 (1S):153-166.
doi: 10.1007/s10584-007-9367-8 URL |
| [4] | Bielenberg D G, Wang Y E, Li Z, Zhebentyayeva T, Fan S, Reighard G L, Scorza R, Abbott A G. 2008. Sequencing and annotation of the evergrowing locus in peach [Prunus persica(L.)Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics & Genomes, 4 (3):495-507. |
| [5] | Campoy J A, Ruiz D, Egea J. 2011. Dormancy in temperate fruit trees in a global warming context:a review. Scientia Horticulturae(Amsterdam), 130 (2):357-372. |
| [6] |
Cooke J E, Eriksson M E, Junttila O. 2012. The dynamic nature of bud dormancy in trees:environmental control and molecular mechanisms. Plant,Cell and Environment, 35 (10):1707-1728.
doi: 10.1111/j.1365-3040.2012.02552.x URL |
| [7] | Corbineau F, Xia Q, Bailly C, El-Maarouf-Bouteau H. 2014. Ethylene,a key factor in the regulation of seed dormancy. Frontiers in Plant Science, 5:539. |
| [8] |
El-Maarouf-Bouteau H, Sajjad Y, Bazin J, Langlade N, Cristescu S M, Balzergue S, Baudouin E, Bailly C. 2015. Reactive oxygen species,abscisic acid and ethylene interact to regulate sunflower seed germination. Plant,Cell and Environment, 38:364-374.
doi: 10.1111/pce.12371 URL |
| [9] |
Feehan J, Harley M, Minnen J V. 2009. Climate change in Europe. 1. Impact on terrestrial ecosystems and biodiversity. A review. Agronomy for Sustainable Development, 29 (3):409-421.
doi: 10.1051/agro:2008066 URL |
| [10] |
Fu J R, Yang S F. 1983. Release of heat pretreatment-induced dormancy in lettuce seeds by ethylene or cytokinin in relation to the production of ethylene and the synthesis of 1-aminocyclopropane-1-carboxylic acid during germination. Journal of Plant Growth Regulation, 2 (1-4):185.
doi: 10.1007/BF02042229 URL |
| [11] |
Horvath D P, Anderson J V, Chao W S, Foley M E. 2003. Knowing when to grow:signals regulating bud dormancy. Trends in Plant Science, 8 (11):534-540.
pmid: 14607098 |
| [12] |
Horvath D P, Sung S, Kim D, Chao W, Anderson J. 2010. Characterization,expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Molecular Biology, 73 (1-2):169-179.
doi: 10.1007/s11103-009-9596-5 pmid: 20066557 |
| [13] | Howe G T, Horvath D P, Dharmawardhana P, Priest H D, Mockler T C, Strauss S H. 2015. Extensive Transcriptome changes during natural onset and release of vegetative bud dormancy in Populus. Frontiers in Plant Science, 6:989. |
| [14] |
Irena Sińska. 1989. Interaction of ethephon with cytokinin and gibberellin during the removal of apple seed dormancy and germination of embryos. Plant Science, 64 (1):39-44.
doi: 10.1016/0168-9452(89)90149-0 URL |
| [15] | Kilany A E, Hegazi E S, Kilany O A. 1990. Ethylene evolution,CO2 product-ion and some endogenous compounds in relation to bud dormancy of pear trees. Bulletin of Faculty of Agriculture, 38 (2):357-367. |
| [16] |
Kucera B, Cohn M A, Leubner-Metzger G. 2005. Plant hormone interactions during seed dormancy release and germination. Seed Science Research, 15 (4):281-307.
doi: 10.1079/SSR2005218 URL |
| [17] |
Lang G A, Early J D, Martin G C, Daenell R L. 1987. Endo-,para-,and ecodormancy:physiological terminology and classification for dormancy research. HortScience, 22 (3):371-377.
doi: 10.21273/HORTSCI.22.3.371 URL |
| [18] |
Li J, Xu Y, Niu Q, He L, Teng Y, Bai S. 2018. Abscisic acid(ABA)promotes the induction and maintenance of pear(Pyrus pyrifolia white pear group)flower bud endodormancy. International Journal of Molecular Sciences, 19 (1):310.
doi: 10.3390/ijms19010310 URL |
| [19] | Li Jian-zhao. 2019. Studies on the molecular mechanism of ABA and related phytohormones-regulated pear bud dormancy[Ph. D. Dissertation]. Hangzhou: Zhejiang University. (in Chinese) |
| 李建召. 2019. 脱落酸及相关激素调控梨芽休眠的分子机制研究[博士论文]. 杭州: 浙江大学. | |
| [20] |
Li P, Zheng T, Zhuo X, Zhang M, Yong X, Li L, Wang J, Cheng T, Zhang Q. 2021. Photoperiod- and temperature-mediated control of the ethylene response and winter dormancy induction in Prunus mume. Horticultural Plant Journal, 7 (3):232-242.
doi: 10.1016/j.hpj.2021.03.005 URL |
| [21] |
Linkies A, Muller K, Morris K, Turečková V, Wenk M, Cadman C S C, Corbineau F, Strnad M, Lynn J R, Finch-Savage W E, Leubner-Metzger G. 2009. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination:a comparative approach using Lepidium sativum and Arabidopsis thaliana. The Plant Cell, 21 (12):3803-3822.
doi: 10.1105/tpc.109.070201 pmid: 20023197 |
| [22] | Niu Qing-feng. 2016. The molecular mechanism and molecular network of pear flower bud dormancy transition[Ph. D. Dissertation]. Hangzhou: Zhejiang University. (in Chinese) |
| 牛庆丰. 2016. 梨花芽休眠转换的分子网络及调控机制[博士论文]. 杭州: 浙江大学. | |
| [23] |
Ophir R, Pang X, Halaly T, Venkateswari J, Lavee S, Galbraith D, Or E. 2009. Gene-expression profiling of grape bud response to two alternative dormancy-release stimuli expose possible links between impaired mitochondrial activity,hypoxia,ethylene-ABA interplay and cell enlargement. Plant Molecular Biology, 71 (4-5):403-423.
doi: 10.1007/s11103-009-9531-9 pmid: 19653104 |
| [24] |
Porto D D, Falavigna V d S, Arenhart R A, Perini P, Buffon V, Anzanello R, dos Santos H P, Fialho F B, Dias de Oliveira P R, Revers L F. 2016. Structural genomics and transcriptional characterization of the Dormancy-Associated MADS-box genes during bud dormancy progression in apple. Tree Genetics & Genomes, 12 (3). Doi: 10.1007/s11295-016-1001-3.
doi: 10.1007/s11295-016-1001-3 URL |
| [25] | Qin Li-jin, Xu Zhen-jun, Yuan Shu-xiang. 2007. Effects of seed soaking with ethephon on seed germination and seedling growth of cucumber in greenhouse in winter. Journal of Anhui Agricultural Sciences,(33):10601-10602. (in Chinese) |
| 秦立金, 徐振军, 袁树祥. 2007. 乙烯利浸种对冬季日光温室黄瓜种子萌发与幼苗生长的影响. 安徽农业科学,(33):10601-10602. | |
| [26] |
Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. 2007. Gene expression during the induction,maintenance,and release of dormancy in apical buds of poplar. Journal of Experimental Botany, 58 (15-16):4047-4060.
doi: 10.1093/jxb/erm261 URL |
| [27] |
Ruonala R, Rinne P L H, Baghour M, Moritz T, Tuominen H, Kangasjarvi J. 2006. Transitions in the functioning of the shoot apical meristem in birch(Betula pendula)involve ethylene. Plant Journal, 46 (4):628-640.
pmid: 16640599 |
| [28] |
Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao R P, Boerjan W, Rohde A. 2007. A molecular timetable for apical bud formation and dormancy induction in poplar. The Plant Cell Online, 19 (8):2370-2390.
doi: 10.1105/tpc.107.052811 URL |
| [29] |
Samish M R. 1954. Dormancy in woody plants. Annual Review of Plant Physiology, 5 (1):183-204.
doi: 10.1146/annurev.pp.05.060154.001151 URL |
| [30] |
Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R. 2011. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiology, 157 (1):485-497.
doi: 10.1104/pp.111.181982 pmid: 21795580 |
| [31] | Shao Xu. 2016. Cloning and functional analysis of dormancy-associated transcription factor(MADS-box)from Chinese cherry flower buds[M. D. Dissertation]. Hangzhou: Zhejiang Normal University. (in Chinese) |
| 邵姁. 2016. 中国樱桃花芽休眠相关MADS-box转录因子克隆与功能分析[硕士论文]. 杭州: 浙江师范大学. | |
| [32] |
Shi Z, Halaly-Basha T, Zheng C, Weissberg M, Ophir R, Galbraith D W, Pang X, Or E. 2018. Transient induction of a subset of ethylene biosynthesis genes is potentially involved in regulation of grapevine bud dormancy release. Plant Molecular Biology, 98 (6):507-523.
doi: 10.1007/s11103-018-0793-y pmid: 30392158 |
| [33] |
Singh R K, Maurya J P, Azeez A, Miskolczi P, Tylewicz S, Stojkovič K, Delhomme N, Busov V, Bhalerao R P. 2018. A genetic network mediating the control of bud break in hybrid aspen. Nature Communications, 9:4173.
doi: 10.1038/s41467-018-06696-y pmid: 30301891 |
| [34] | Subbiah V, Reddy K J. 2010. Interactions between ethylene,abscisic acid and cytokinin during germination and seedling establishment in Arabidopsis. Journal of Bioscience and Bioengineering, 35 (3):451-458. |
| [35] |
Song Song-quan, Liu Jun, Xu Heng-heng, Zhang Qi, Huang Hui, Wu Xian-jin. 2019. Biosynthesis and signaling of ethylene and their regulation on seed germination and dormancy. Acta Agronomica Sinica, 45 (7):969-981. (in Chinese)
doi: 10.3724/SP.J.1006.2019.84175 |
|
宋松泉, 刘军, 徐恒恒, 张琪, 黄荟, 伍贤进. 2019. 乙烯的生物合成与信号及其对种子萌发和休眠的调控. 作物学报, 45 (7):969-981.
doi: 10.3724/SP.J.1006.2019.84175 |
|
| [36] | Tian Wen-jie. 2018. Effect of ethephon soaking on seed germination of maize. Modern Agricultural Science and Technology,(15):1-2. (in Chinese) |
| 田文杰. 2018. 乙烯利浸种对玉米种子萌发的影响. 现代农业科技,(15):1-2. | |
| [37] |
Ubi B E, Sakamoto D, Ban Y, Shimada T, Ito A, Nakajima I, Takemura Y, Tamura F, Saito T, Moriguchi T. 2010. Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. Journal of the American Society for Horticultural Science, 135 (2):174-182.
doi: 10.21273/JASHS.135.2.174 URL |
| [38] |
Wu R M, Walton E F, Richardson A C, Wood M, Hellens R P, Varkonyi-Gasic E. 2012. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. Journal of Experimental Botany, 63 (2):797-807.
doi: 10.1093/jxb/err304 URL |
| [39] |
Wu R, Wang T, McGie T, Voogd C, Allan A C, Hellens R P, Varkonyi-Gasic E. 2014. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals,but has no effect on vegetative growth,dormancy,or flowering time. Journal of Experimental Botany, 65 (17):4985-4995.
doi: 10.1093/jxb/eru264 URL |
| [40] |
Wu R, Wang T, Warren B A W, Allan A C, Macknight R C, Varkonyi-Gasic E. 2017. Kiwifruit SVP2 gene prevents premature budbreak during dormancy. Journal of Experimental Botany, 68 (5):1071-1082.
doi: 10.1093/jxb/erx014 URL |
| [41] |
Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. 2008. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. Journal of the American Society for Horticultural Science, 133 (5):708-716.
doi: 10.21273/JASHS.133.5.708 URL |
| [42] | Yan Xin-hui. Identification and functional analysis of pear dormancy associated DAM genes[M. D. Dissertation]. Hangzhou: Zhejiang University. (in Chinese) |
| 鄢馨卉. 2019. 梨休眠相关DAM基因的鉴定及功能分析[硕士论文]. 杭州: 浙江大学. | |
| [43] |
Yang Q, Niu Q, Li J, Zheng X, Ma Y, Bai S, Teng Y. 2018. PpHB22,a member of HD-Zip proteins,activates,PpDAM1,to regulate bud dormancy transition in'Suli'pear(Pyrus pyrifolia white pear group). Plant Physiology and Biochemistry, 127:355-365.
doi: 10.1016/j.plaphy.2018.04.002 URL |
| [44] |
Yang Q, Yang B, Li J, Wang Y, Tao R, Yang F, Wu X, Yan X, Ahmad M, Shen J, Bai S, Teng Y. 2020. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant,Cell and Environment, 43 (6):1360-1375.
doi: 10.1111/pce.13744 URL |
| [45] |
Zheng C, Acheampong A K, Shi Z, Mugzech A, Halaly-Basha T, Shaya F, Sun Y, Colova V, Mosquna A, Ophir R, Galbraith D W, Or E. 2018. Abscisic acid catabolism enhances dormancy release of grapevine buds. Plant,Cell and Environment, 41 (10):2490-2503.
doi: 10.1111/pce.13371 URL |
| [1] | SONG Jiankun, YANG Yingjie, LI Dingli, MA Chunhui, WANG Caihong, and WANG Ran. A New Pear Cultivar‘Luxiu’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 3-4. |
| [2] | DONG Xingguang, CAO Yufen, ZHANG Ying, TIAN Luming, HUO Hongliang, QI Dan, XU Jiayu, LIU Chao, and WANG Lidong. A New Cold-resistant Crispy Pear Cultivar‘Yucuixiang’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 5-6. |
| [3] | OU Chunqing, JIANG Shuling, WANG Fei, MA Li, ZHANG Yanjie, and LIU Zhenjie. A New Early-ripening Pear Cultivar‘Xingli Mishui’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 7-8. |
| [4] | ZHANG Yanjie, WANG Fei, OU Chunqing, MA Li, JIANG Shuling, and LIU Zhenjie. A New Pear Cultivar‘Zhongli Yucui 3’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 9-10. |
| [5] | WANG Suke, LI Xiugen, YANG Jian, WANG Long, SU Yanli, ZHANG Xiangzhan, and XUE Huabai. A New Red Pear Cultivar‘Danxiahong’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 13-14. |
| [6] | LÜ Yi, SUI Jing, XUE Yuping, HUANG Haijing, MENG Yanling, WANG Tongyong, and YANG He, . A New Fig Cultivar‘Jinqing’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 45-46. |
| [7] | WANG Fei, OU Chunqing, ZHANG Yanjie, MA Li, and JIANG Shuling. A New Late Ripening Pear Cultivar‘Huaqiu’with Long Storage Period [J]. Acta Horticulturae Sinica, 2022, 49(S1): 9-10. |
| [8] | SONG Jiankun, LI Dingli, YANG Yingjie, MA Chunhui, WANG Caihong, and WANG Ran. A New Pear Cultivar‘Qindaohong’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 11-12. |
| [9] | MA Ke, HU Shan, WANG Yan, LI Jiaqiang, LAI Yongchao, and MA Huiqin, . A New Fig Cultivar‘Caiyi’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 31-32. |
| [10] | ZHU Shiping, WEN Rongzhong, WANG Yuanyuan, and ZENG Yang. A New Very Late Ripening Citrus Variety‘Jinlegan’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 43-44. |
| [11] | JIN Jianpeng, GAO Jie, XIE Qi, WEI Yonglu, LU Chuqiao, YANG Fengxi, and ZHU Genfa. A New Cymbidium sinense Cultivar‘Sijihua’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 127-128. |
| [12] | LI Dongmei, ZHAN Qicheng, HUANG Dan, JIANG Xionghui, YU Bo, LI Dongjun, ZHANG Hua, WANG Zhen, CAI Guiqi, and ZHU Genfa, . A New Aglaonema Cultivar‘Hongjian’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 155-156. |
| [13] | LI Dongmei, ZHAN Qicheng, HUANG Dan, JIANG Xionghui, YU Bo, LI Dongjun, CAI Guiqi, WANG Zhen, ZHANG Hua, and ZHU Genfa, . A New Aglaonema Cultivar‘Baibaoshi’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 155-156. |
| [14] | GUO Weizhen, ZHAO Jingxian, LI Ying. A New Mid-early Ripening Pear Cultivar‘Meiyu’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 2051-2052. |
| [15] | LIU Jinming, GUO Caihua, YUAN Xing, KANG Chao, QUAN Shaowen, NIU Jianxin. Genome-wide Identification of Dof Family Genes and Expression Analysis Sepal Persistent and Abscission in Pear [J]. Acta Horticulturae Sinica, 2022, 49(8): 1637-1649. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd