
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (9): 2343-2352.doi: 10.16420/j.issn.0513-353x.2024-0944
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
SHENG Xiaoguang1, YU Huifang1, WANG Jiansheng1, SHEN Yusen1, SONG Mengfei1, QIAO Shuting2, LIN Fan3, DU Sifan4, LI Jiaojiao3, GU Honghui1,*( )
)
Received:2025-03-20
Revised:2025-05-26
Online:2025-09-25
Published:2025-09-24
Contact:
GU Honghui
SHENG Xiaoguang, YU Huifang, WANG Jiansheng, SHEN Yusen, SONG Mengfei, QIAO Shuting, LIN Fan, DU Sifan, LI Jiaojiao, GU Honghui. Double Populations Genetic Mapping of the Main QTL Locus qDCA.C8-1 for the Curd Appearance Trait of Cauliflower[J]. Acta Horticulturae Sinica, 2025, 52(9): 2343-2352.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0944

Fig. 1 Phenotypic distribution of curd appearance trait in parents of C4101,J1401,R06 and their constructed double F2 populations A:Curd appearance phenotype of female parent(cauliflower C4101),male parents of Chinese kale(J1401)and broccoli landrace(R06);B,C:CJF2 population and its parents;D,E:CRF2 population and its parents
| 群体 Populations | 染色体 Chromosomes | QTL位点 QTL locus | 位置/cM Position | 阈值 LOD | 加性效应 Additive effect | 贡献率/% R2 |
|---|---|---|---|---|---|---|
| CJF2 | 1 | 1 | 1 198.01 | 11.09 | -4.71 | 15.21 |
| 2 | 2 | 1 018.61 | 4.61 | 2.39 | 4.40 | |
| 7 | 3 | 790.91 | 3.07 | 1.81 | 2.50 | |
| 8 | 4 | 522.61 | 9.42 | 3.52 | 8.95 | |
| CRF2 | 1 | 1 | 609.11 | 2.79 | -2.88 | 3.05 |
| 3 | 2 | 696.31 | 2.85 | -2.87 | 3.11 | |
| 4 | 3 | 7.31 | 5.04 | 3.24 | 4.70 | |
| 4 | 4 | 219.61 | 3.59 | 2.93 | 3.38 | |
| 5 | 5 | 237.81 | 6.17 | -3.59 | 7.10 | |
| 6 | 6 | 760.31 | 3.93 | 2.90 | 3.69 | |
| 7 | 7 | 562.81 | 3.05 | 4.31 | 4.03 | |
| 8 | 8 | 567.11 | 3.08 | -2.36 | 3.01 | |
| 8 | 9 | 878.11 | 5.88 | 3.63 | 6.43 | |
| 9 | 10 | 898.01 | 3.15 | -2.76 | 2.98 |
Table 1 QTLs associated with curd appearance trait of cauliflower
| 群体 Populations | 染色体 Chromosomes | QTL位点 QTL locus | 位置/cM Position | 阈值 LOD | 加性效应 Additive effect | 贡献率/% R2 |
|---|---|---|---|---|---|---|
| CJF2 | 1 | 1 | 1 198.01 | 11.09 | -4.71 | 15.21 |
| 2 | 2 | 1 018.61 | 4.61 | 2.39 | 4.40 | |
| 7 | 3 | 790.91 | 3.07 | 1.81 | 2.50 | |
| 8 | 4 | 522.61 | 9.42 | 3.52 | 8.95 | |
| CRF2 | 1 | 1 | 609.11 | 2.79 | -2.88 | 3.05 |
| 3 | 2 | 696.31 | 2.85 | -2.87 | 3.11 | |
| 4 | 3 | 7.31 | 5.04 | 3.24 | 4.70 | |
| 4 | 4 | 219.61 | 3.59 | 2.93 | 3.38 | |
| 5 | 5 | 237.81 | 6.17 | -3.59 | 7.10 | |
| 6 | 6 | 760.31 | 3.93 | 2.90 | 3.69 | |
| 7 | 7 | 562.81 | 3.05 | 4.31 | 4.03 | |
| 8 | 8 | 567.11 | 3.08 | -2.36 | 3.01 | |
| 8 | 9 | 878.11 | 5.88 | 3.63 | 6.43 | |
| 9 | 10 | 898.01 | 3.15 | -2.76 | 2.98 |
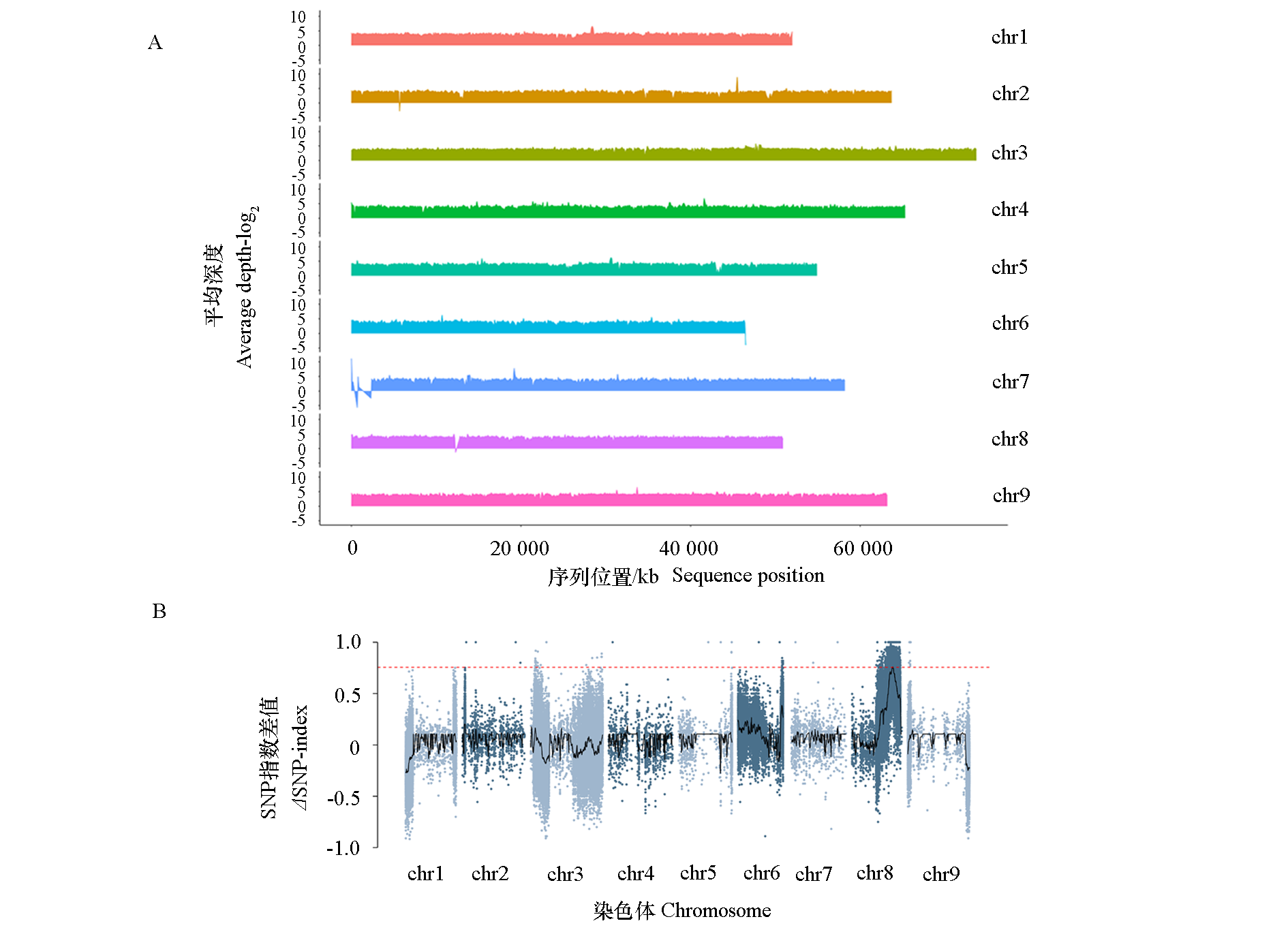
Fig. 2 BSA analysis of extreme individuals with curd appearance trait in cauliflower F2:3 population A:Parent genome SNP for BSA analysis;B:Δ(SNP-index)plots of F2:3 BSA-seq results
| 引物名称 Primers | 变异位点 Variation locus | 序列信息(5′-3′) Sequencing information |
|---|---|---|
| FC84287 | T/A | FHEX:GAAGGTGACCAAGTTCATGCTTATGGAAATCTGCTAGTTT |
| FFAM:GAAGGTCGGAGTCAACGGATTTATGGAAATCTGCTAGTTA | ||
| R:TCGTATATATCCACTCCGAACT | ||
| RC84556 | C/A | FHEX:GAAGGTGACCAAGTTCATGCTCAGCCACGGATGTAAAGTCTTCC |
| FFAM:GAAGGTCGGAGTCAACGGATTCAGCCACGGATGTAAAGTCTTCA | ||
| R:TAGAACACGCTACTTGGTTGCAGG |
Table 2 Primer sequence information for KASP markers FC84287 and RC84556
| 引物名称 Primers | 变异位点 Variation locus | 序列信息(5′-3′) Sequencing information |
|---|---|---|
| FC84287 | T/A | FHEX:GAAGGTGACCAAGTTCATGCTTATGGAAATCTGCTAGTTT |
| FFAM:GAAGGTCGGAGTCAACGGATTTATGGAAATCTGCTAGTTA | ||
| R:TCGTATATATCCACTCCGAACT | ||
| RC84556 | C/A | FHEX:GAAGGTGACCAAGTTCATGCTCAGCCACGGATGTAAAGTCTTCC |
| FFAM:GAAGGTCGGAGTCAACGGATTCAGCCACGGATGTAAAGTCTTCA | ||
| R:TAGAACACGCTACTTGGTTGCAGG |
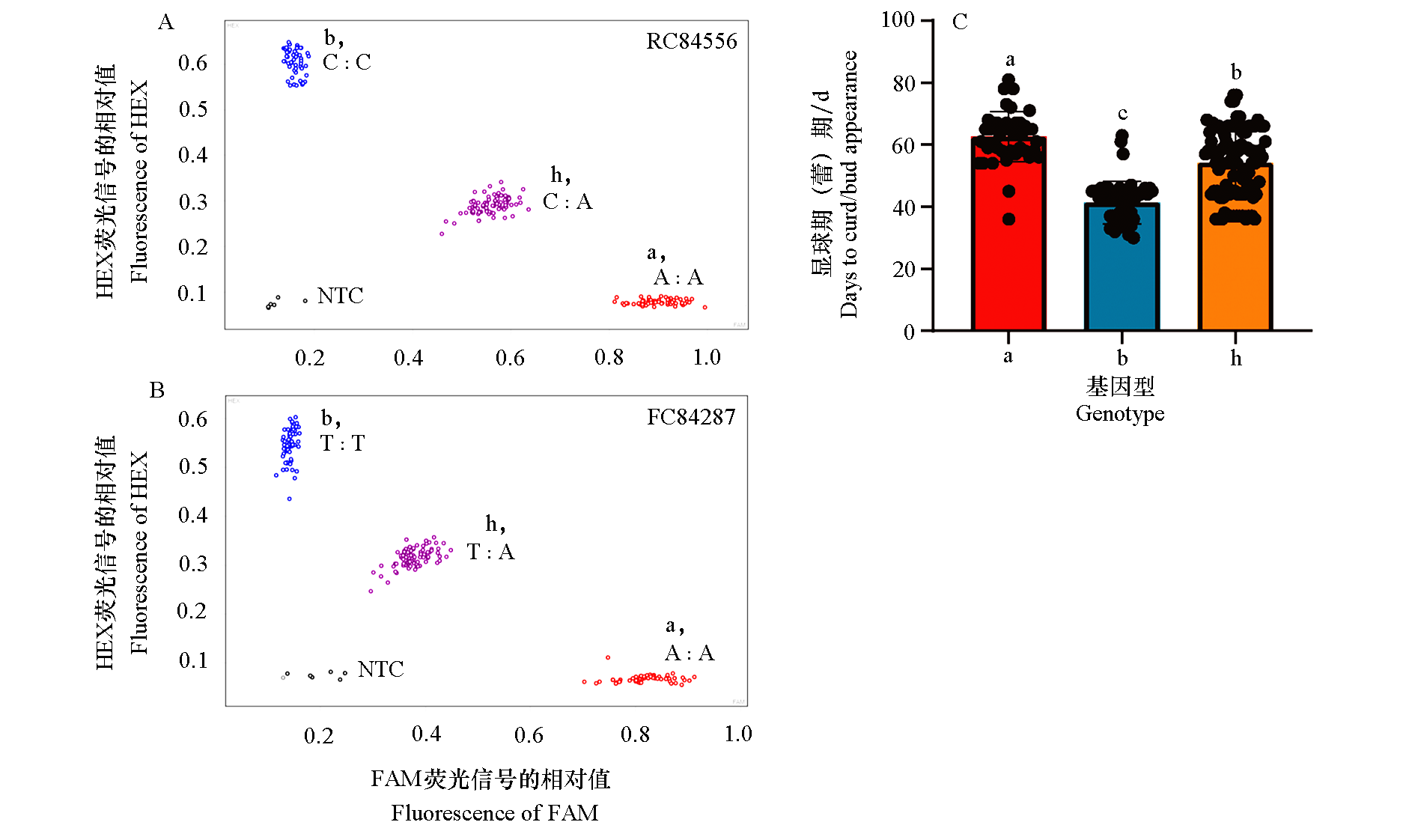
Fig. 3 KASP genotyping of F2:3 population using markers FC84287 and RC84556 A,B:FC84287 and RC84556 markers genotyping result;C:Distribution of curd appearance trait among three genotypes(a:C4101 genotype;b:J1401genotype;h:Heterozygous genotype). NTC:H2O,black control. Different lowercase letters indicate significant differences at 0.05 level
| [1] |
doi: 10.1126/science.abg5999 pmid: 34244409 |
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
doi: 10.1038/s41588-024-01744-4 pmid: 38714866 |
| [6] |
doi: 10.1104/pp.110.164160 pmid: 20855520 |
| [7] |
doi: 10.11924/j.issn.1000-6850.casb2024-0032 |
|
高婧, 秦梦凡, 李坤, 李武. 2024. 玉米产量性状遗传特性研究进展的可视化分析. 中国农学通报, 40 (16):156-164.
doi: 10.11924/j.issn.1000-6850.casb2024-0032 |
|
| [8] |
|
| [9] |
|
| [10] |
|
|
贾俊忠, 田丽萍, 薛琳, 魏亦农. 2010. 番茄可溶性固形物的动态QTL及相关性状. 遗传, 32 (10):1077-1083.
|
|
| [11] |
doi: 10.1126/science.7824951 pmid: 7824951 |
| [12] |
|
| [13] |
|
|
李锡香, 方智远. 2008. 花椰菜和青花菜种质资源描述规范和数据标准. 北京: 中国农业出版社:58-60.
|
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
doi: 10.16420/j.issn.0513-353x.2025-0177 |
|
孙德岭, 姚星伟, 陈锐, 杨迎霞, 单晓政. 2025. 国花椰菜育种50 年回顾与进展. 园艺学报, 52 (5):1159-1179.
doi: 10.16420/j.issn.0513-353x.2025-0177 |
|
| [20] |
doi: 10.1007/s00122-019-03466-2 pmid: 31676958 |
| [21] |
|
| [22] |
doi: 10.16420/j.issn.0513-353x.2023-0946 |
|
文宋琴, 李佳霖, 池卓恒, 夏燕, 王淑明, 吴頔, 郝雅雯, 郭启高, 梁国鲁, 景丹龙. 2024. 光和温度对开花时间基因可变剪接的影响研究进展. 园艺学报, 51 (8):1949-1963.
|
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
|
俎峰, 赵凯琴, 张云云, 田正书, 刘亚俊, 奚俊玉, 束正齐, 符明联. 2019. 甘蓝型油菜的花期与生育期QTL定位. 南方农业学报, 50 (3):500-505.
|
| [1] | ZHU Shiyang, LIU Qing, ZHONG Weijie, and TANG Zheng. A New Cultivar of Cauliflower‘Wenxuan70’ [J]. Acta Horticulturae Sinica, 2025, 52(7): 1937-1938. |
| [2] | SUN Deling, YAO Xingwei, CHEN Rui, YANG Yingxia, SHAN Xiaozheng. Review and Prospects of Cauliflower Breeding During the Past 50 Years in China [J]. Acta Horticulturae Sinica, 2025, 52(5): 1159-1179. |
| [3] | WANG Qingbiao, WANG Yanping, WU Xiangyu, CHEN Ningjie, CHI Ting, HUA Dingzhu, GE Ailing, GUO Yu, WEI Lei, ZHANG Li. Genetic Analysis and Preliminary Mapping of the Fertility Restoration Gene for NWB-CMS in Radish(Raphanus sativus) [J]. Acta Horticulturae Sinica, 2025, 52(5): 1351-1363. |
| [4] | ZHANG Yiying, ZHANG Yu, CHU Yunxia, CHEN Hairong, DENG Shan, LI Aiai, ZHAO Hong, LIU Kun, HAN Ruixi, REN Li. Color Characteristics of Broccoli and Cauliflower Leaves Based on the Colorimeter Parameters [J]. Acta Horticulturae Sinica, 2025, 52(4): 872-882. |
| [5] | XIA Zhilei, YU Bingxin, YANG Meng, LIAN Zilin, GUAN Lilan, HE Yichao, YAN Shuangshuang, CAO Bihao, QIU Zhengkun. Fine-Mapping and KASP Marker Development for the Gene Underlying Green Fruit in Eggplant [J]. Acta Horticulturae Sinica, 2024, 51(9): 2008-2018. |
| [6] | LI Ken, ZHANG Wei, WU Yunpeng, PENG Dongxiu, ZHANG Ruowei. Development and Utilization of KASP Marker for Identification of Pulp Firmness in Melon [J]. Acta Horticulturae Sinica, 2024, 51(4): 773-786. |
| [7] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, YAO Xingwei. Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| [8] | QIAO Jun, LIU Jing, LI Suwen, WANG Liying. Prediction of Fruit Color Genes Under the Calyx of Eggplant Based on Genome-wide Resequencing in an Extreme Mixing Pool [J]. Acta Horticulturae Sinica, 2022, 49(3): 613-621. |
| [9] | SHENG Yunyan, YANG Limin, DAI Dongyang, ZHANG Jiaxin, WANG Ling, WANG Di, CAI Yi, TIAN Limei. Cytological Observation of Fruit Peduncle Abscission Zone and Preliminary Mapping of Mature Fruit Abscission AL3 gene in Melon [J]. Acta Horticulturae Sinica, 2022, 49(2): 341-351. |
| [10] | YANG Shuangjuan, Zhang Xiaowei, WEI Xiaochun, ZHAO Yanyan, WANG Zhiyong, ZHAO Xiaobin, LI Lin, YUAN Yuxiang. Mapping and KASP Markers Development for Clubroot Resistance Gene BraA.Pb.8.4 in Chinese Cabbage [J]. Acta Horticulturae Sinica, 2021, 48(7): 1317-1328. |
| [11] | CHENG Feng, SONG Mengfei, CAO Lei, ZHANG Mengru, YANG Zhige, CHEN Jinfeng, LOU Qunfeng. Genetic Mapping for a Medium Short-fruit Mutant of Cucumber [J]. Acta Horticulturae Sinica, 2021, 48(7): 1359-1370. |
| [12] | ZHU Shiyang, ZHANG Xiaoling, LIU Qing, ZHONG Weijie, LUO Tiankuan, TANG Zheng, XU Qian, ZHOU Yuanchang. Cytoplasmic Effect Analysis of Different CMS Sources of Cauliflower [J]. Acta Horticulturae Sinica, 2021, 48(6): 1107-1122. |
| [13] | YU Hailong, REN Wenjing, FANG Zhiyuan, YANG Limei, ZHUANG Mu, LÜ Honghao, WANG Yong, JI Jialei, ZHANG Yangyong. Advances on Fertility Restoration for Cytoplasmic Male Sterility in Vegetables [J]. Acta Horticulturae Sinica, 2021, 48(5): 1031-1046. |
| [14] | ZHANG Hui, ZHANG Shujiang, LI Fei, ZHANG Shifan, LI Guoliang, MA Xiaochao, LIU Xitong, and SUN Rifei. Research Progress on Clubroot Disease Resistance Breeding of Brassica rapa [J]. Acta Horticulturae Sinica, 2020, 47(9): 1648-1662. |
| [15] | GAO Peng, LIU Shi, CUI Haonan, ZHANG Taifeng, WANG Xuezheng, LIU Hongyu, ZHU Zicheng, and LUAN Feishi. Research Progress of Melon Genomics,Functional Gene Mapping and Genetic Engineering [J]. Acta Horticulturae Sinica, 2020, 47(9): 1827-1844. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd