
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (9): 2377-2386.doi: 10.16420/j.issn.0513-353x.2024-0842
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
LI Maofu1,2,3, YANG Yuan1,2,4, WANG Hua1,2,3, SUN Pei1,2,3, KANG Yanhui1,2,3, SUN Xiangyi1,2,3, LUO Jie1,2,3, LIU Xin1,2,3, JIN Min1,2,3, JIN Wanmei1,2,3,*( )
)
Received:2025-01-05
Revised:2025-04-07
Online:2025-09-25
Published:2025-09-24
Contact:
JIN Wanmei
LI Maofu, YANG Yuan, WANG Hua, SUN Pei, KANG Yanhui, SUN Xiangyi, LUO Jie, LIU Xin, JIN Min, JIN Wanmei. Cloning and Preliminary Functional Exploration of the Transposon-Derived Gene RcHTD in Rose[J]. Acta Horticulturae Sinica, 2025, 52(9): 2377-2386.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0842
| 目的Purpose | 引物名称Primer name | 引物序列(5′-3′)Primer sequence |
|---|---|---|
| 基因克隆 Gene cloning | RcHTD | F:ATGGAAGTCACCCCTTTCACATTTCT |
| R:CTAATGAAATCCAGTGCCTGCAAGGCC | ||
| RT-PCR | RcHTD | F:GCGGAGGAAGGTCGATGAGGACGATACC |
| R:CCTGTAATTCGCGTCAAGAGTCTCC | ||
| Actin | F:GGCTGTTCTCTCTCTGTATGC | |
| R:TTCTGGGCACCTGAATCTC | ||
| 亚细胞定位 Subcellular localization | RcHTD | F: CGACTCTAGTCTAGAAAGCTATGGAAGTCACCCCTTTCACATTTCT |
| R: CATGGTACCGGATCCACTAGTATGAAATCCAGTGCCTGCAAGGCC | ||
| 酵母双杂交 Yeast two hybrid | RcHTD | F: TGGCCATGGAGGCCAGTGAATTCATGGAAGTCACCCCTTTCACATTTCT |
| R: CTACGATTCATCTGCAGCTCGAGCTAATGAAATCCAGTGCCTGCAAGGCC | ||
| 蛋白诱导表达 Protein induced expression EMAS 探针 EMAS probe | RcHTD Rosa1 | F: GATCTGGTTCCGCGTGGATCCATGGAAGTCACCCCTTTCACATTTCT |
| R: GATGCGGCCGCTCGAGTCGAATGAAATCCAGTGCCTGCAAGGCC AAATGAATGCTTACTTTCGGAAGTCCTTGTTC AAAAAAAAAAAAAAAATCGGAAGTCCTTGTTC |
Table 1 Primer sequence used in this study
| 目的Purpose | 引物名称Primer name | 引物序列(5′-3′)Primer sequence |
|---|---|---|
| 基因克隆 Gene cloning | RcHTD | F:ATGGAAGTCACCCCTTTCACATTTCT |
| R:CTAATGAAATCCAGTGCCTGCAAGGCC | ||
| RT-PCR | RcHTD | F:GCGGAGGAAGGTCGATGAGGACGATACC |
| R:CCTGTAATTCGCGTCAAGAGTCTCC | ||
| Actin | F:GGCTGTTCTCTCTCTGTATGC | |
| R:TTCTGGGCACCTGAATCTC | ||
| 亚细胞定位 Subcellular localization | RcHTD | F: CGACTCTAGTCTAGAAAGCTATGGAAGTCACCCCTTTCACATTTCT |
| R: CATGGTACCGGATCCACTAGTATGAAATCCAGTGCCTGCAAGGCC | ||
| 酵母双杂交 Yeast two hybrid | RcHTD | F: TGGCCATGGAGGCCAGTGAATTCATGGAAGTCACCCCTTTCACATTTCT |
| R: CTACGATTCATCTGCAGCTCGAGCTAATGAAATCCAGTGCCTGCAAGGCC | ||
| 蛋白诱导表达 Protein induced expression EMAS 探针 EMAS probe | RcHTD Rosa1 | F: GATCTGGTTCCGCGTGGATCCATGGAAGTCACCCCTTTCACATTTCT |
| R: GATGCGGCCGCTCGAGTCGAATGAAATCCAGTGCCTGCAAGGCC AAATGAATGCTTACTTTCGGAAGTCCTTGTTC AAAAAAAAAAAAAAAATCGGAAGTCCTTGTTC |
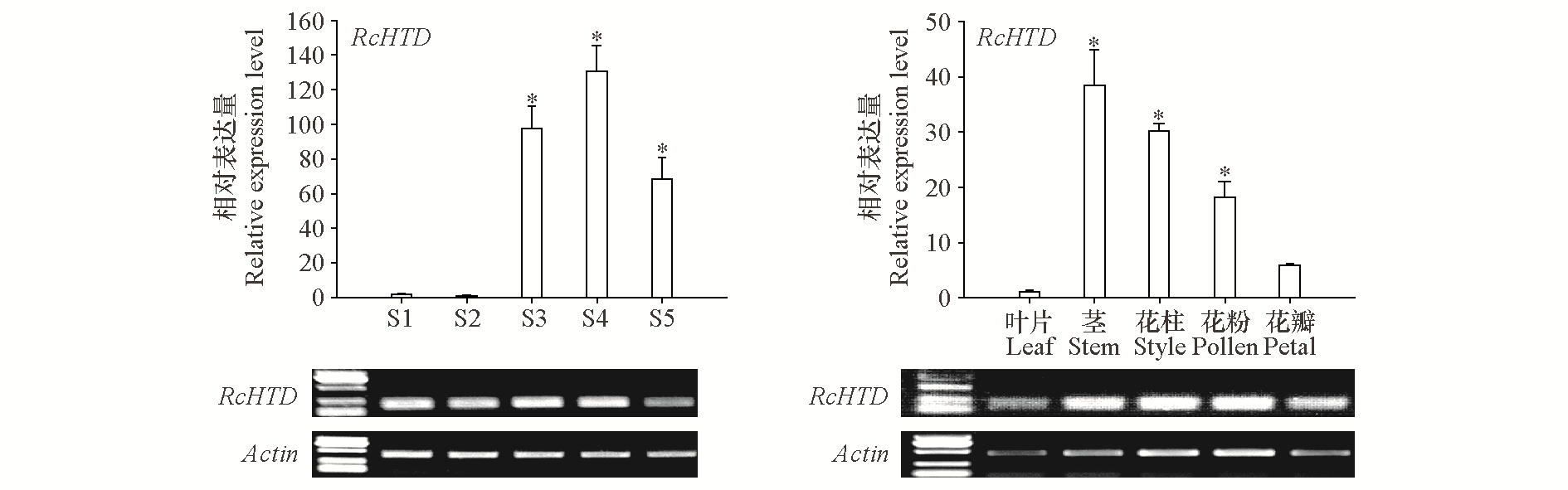
Fig. 5 Expression analysis of RcHTD gene in different developmental stages and tissues of Rosa chinensis‘Slater’s Crimson China’petals S1:Small bud stage;S2:Big bud stage;S3:Initial opening period;S4:Blooming period;S5:Withering period. t-Test,*P < 0.05
| [1] |
doi: 10.1186/s13059-018-1577-z pmid: 30454069 |
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
doi: 10.1038/nrg793 pmid: 11988759 |
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
|
李茂福, 杨媛, 王华, 刘佳棽, 金万梅. 2017. 月季bHLH基因的克隆、表达及其与MYB和WD40的互作分析. 园艺学报, 44 (10):1949-1958.
doi: 10.16420/j.issn.0513-353x.2017-0183 |
|
| [14] |
|
|
刘珂珂, 汤定钦, 周明兵. 2021. 转座子衍生基因的演变途径及对生物生长发育的影响. 中国细胞生物学学报, 43 (1):152-160
|
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
doi: S0092-8674(18)30843-2 pmid: 30057117 |
| [25] |
doi: 10.1038/s41588-022-01214-9 pmid: 36396707 |
| [26] |
doi: 10.16420/j.issn.0513-353x.2020-0361 |
|
相元萍, 王一丹, 贺洪军, 徐启江. 2020. 植物转座子类型及其插入突变对园艺植物花发育影响的研究进展. 园艺学报, 47 (11):2247-2266.
|
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [1] | MO Jinxia, LI Fang, XIONG Xinting, ZHONG Zaofa, PENG Ting. Characterization of PtSARD1 and Its Preliminary Role in Drought Tolerance [J]. Acta Horticulturae Sinica, 2025, 52(6): 1399-1411. |
| [2] | MA Yuwan, LIU Ao, LIU Xiangdong, ZHANG Yajing, DONG Xuanke, LI Yufan, CHEN Jiren. Cloning of the RcRAP2.7 Gene and its Expression Analysis Under Abiotic Stresses in Rosa chinensis‘Yueyuehong’ [J]. Acta Horticulturae Sinica, 2025, 52(4): 921-932. |
| [3] | REN Hengze, LI Danying, YU Yating, LÜ Wuyun, HAO Xinyuan, WANG Xinchao, WANG Yuchun. Research Advances of Strategies to Engineer VIGS Vectors and Its Application in Plants [J]. Acta Horticulturae Sinica, 2024, 51(7): 1455-1473. |
| [4] | LU Jing, WEI Suyun, YIN Tongming, CHEN Yingnan. Research Progress on the Cytokinin Type-A Response Regulator Gene Family [J]. Acta Horticulturae Sinica, 2023, 50(9): 1867-1888. |
| [5] | GUO Jing, LIAO Manyu, JIN Yan, MA Xiaochuan, ZHANG Fen, LU Xiaopeng, DENG Ziniu, SHENG Ling. Functional Analysis of Transcription Factor CsbHLH3 in Regulating Citric Acid Metabolism of Citrus Fruit [J]. Acta Horticulturae Sinica, 2023, 50(5): 947-958. |
| [6] | SHEN Yuxiao, ZOU Jinyu, LUO Ping, SHANG Wenqian, LI Yonghua, HE Songlin, WANG Zheng, SHI Liyun. Genome-wide Identification and Abiotic Stress Response Analysis of PP2C Family Genes in Rosa chinensis‘Old Blush’ [J]. Acta Horticulturae Sinica, 2023, 50(10): 2139-2156. |
| [7] | HUANG Ling, HU Xianmei, LIANG Zehui, WANG Yanping, CHAN Zhulong, XIANG Lin. Cloning and Function Identification of Anthocyanidin Synthase Gene TgANS in Tulipa gesneriana [J]. Acta Horticulturae Sinica, 2022, 49(9): 1935-1944. |
| [8] | YANG Yuyan, DUAN Xinyuan, HE Zhilin, BING Qihao, CHEN Suoying, LIU Xiaoman, ZENG Ming, LIU Xiaogang. Cloning and Function Characterization of UDP-L-rhamnose Synthase from Fortunella crassifolia [J]. Acta Horticulturae Sinica, 2022, 49(8): 1663-1672. |
| [9] | TAO Xin, ZHU Rongxiang, GONG Xin, WU Lei, ZHANG Shaoling, ZHAO Jianrong, ZHANG Huping. Fructokinase Gene PpyFRK5 Plays an Important Role in Sucrose Accumulation of Pear Fruit [J]. Acta Horticulturae Sinica, 2022, 49(7): 1429-1440. |
| [10] | MA Mingying, HAO Chenxing, ZHANG Kai, XIAO Guihua, SU Hanying, WEN Kang, DENG Ziniu, MA Xianfeng. CsSWEET2a Promotes the Infection of Xanthomonas citri subsp. citri [J]. Acta Horticulturae Sinica, 2022, 49(6): 1247-1260. |
| [11] | LI Chonghui, YANG Guangsui, ZHANG Zhiqun, YIN Junmei. A Novel R2R3-MYB Transcription Factor Gene AaMYB6 Involved in Anthocyanin Biosynthesis in Anthurium andraeanum [J]. Acta Horticulturae Sinica, 2021, 48(10): 1859-1872. |
| [12] | ZHENG Hao, ZHANG Fen, JIAN Yue, HUANG Wenli, LIANG Sha, JIANG Min, YUAN Qiao, WANG Qiaomei, SUN Bo. Cloning and Function Identification of Dihydroflavonol 4-Reductase Gene BoaDFR in Chinese Kale [J]. Acta Horticulturae Sinica, 2021, 48(1): 73-82. |
| [13] | WU Jinghua,YANG Chao,and WU Shaohua*. Expression and Function Analysis of AGL6 Homologous Genes in Cymbidium ensifolium [J]. ACTA HORTICULTURAE SINICA, 2019, 46(6): 1123-1133. |
| [14] | LIU Shengyu1,ZHANG Shikui2,LU Juanfang1,LI Wenhui2,and XI Wanpeng1,3,*. Molecular Cloning and Function Analysis of Carotenoid Cleavage Dioxygenase 1(CCD1)in Apricot Fruit [J]. ACTA HORTICULTURAE SINICA, 2018, 45(3): 471-482. |
| [15] | ZHANG Shi-Zhong, FU Ying, ZHOU Bo, JIANG Ze-Sheng, XU Rui-Rui, SHU Huai-Rui. The Construction and Instruction Manual of Apple Functional Genomic Database [J]. ACTA HORTICULTURAE SINICA, 2012, 39(11): 2245-2250. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd