
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (6): 1189-1200.doi: 10.16420/j.issn.0513-353x.2023-0500
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
ZHAO Chongbin1, ZHANG Weiyin1, LI Shuqing1, GUO Yihan1, XU Hongxia2, CHEN Junwei2,**( ), YANG Xianghui1,**(
), YANG Xianghui1,**( ), PENG Ze1,**(
), PENG Ze1,**( )
)
Received:2023-09-11
Revised:2023-11-21
Online:2024-12-18
Published:2024-06-21
Contact:
CHEN Junwei, YANG Xianghui, PENG Ze
ZHAO Chongbin, ZHANG Weiyin, LI Shuqing, GUO Yihan, XU Hongxia, CHEN Junwei, YANG Xianghui, PENG Ze. Quantitative Trait Locus Mapping and Analysis of Traits Related to Flower,Fruit,and Leaf in Loquat[J]. Acta Horticulturae Sinica, 2024, 51(6): 1189-1200.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0500
| 叶面形态 Leaf blade | 叶姿 Leaf attitude | 花支轴紧密度 Density of secondary peduncles | 叶形 Leaf shape | 叶基形状 Shape of leaf base | 赋值 Assignment |
|---|---|---|---|---|---|
| 平展Flat | 斜向上Upward | 紧密Dense | 披针形Lanceolate | 渐狭形Narrowly cuneate | 1 |
| 稍皱Rugulose | 平伸Outward | 中等Medium | 椭圆形Elliptic | 楔形Cuneate | 2 |
| 皱Ruguse | 斜向下Downward | 疏散Sparse | 倒卵形Obovoid | 宽楔形Broadly cuneate | 3 |
Table 1 Description and assignment of hypothetical qualitative traits
| 叶面形态 Leaf blade | 叶姿 Leaf attitude | 花支轴紧密度 Density of secondary peduncles | 叶形 Leaf shape | 叶基形状 Shape of leaf base | 赋值 Assignment |
|---|---|---|---|---|---|
| 平展Flat | 斜向上Upward | 紧密Dense | 披针形Lanceolate | 渐狭形Narrowly cuneate | 1 |
| 稍皱Rugulose | 平伸Outward | 中等Medium | 椭圆形Elliptic | 楔形Cuneate | 2 |
| 皱Ruguse | 斜向下Downward | 疏散Sparse | 倒卵形Obovoid | 宽楔形Broadly cuneate | 3 |
| 性状 Trait | 果纵径Longitudinal diameter (LD) | 果横径Transverse diameter (TD) | 果形指数Fruit shape index (FSI) | 果肉厚 Flesh thickness (FT) | 可溶性固形物Total soluble solids (TSS) | 种子质量 Seed weight (SW) | 种子数 Seed number (SN) | 叶姿 Leaf attitude(LA) | |
|---|---|---|---|---|---|---|---|---|---|
| LD | 1 | ||||||||
| TD | 0.735** | 1 | |||||||
| FSI | 0.248** | -0.472** | 1 | ||||||
| FT | 0.766** | 0.822** | -0.178* | 1 | |||||
| TSS | -0.440** | -0.496** | 0.136 | -0.493** | 1 | ||||
| SW | 0.568** | 0.894** | -0.532** | 0.517** | -0.322** | 1 | |||
| SN | 0.016 | 0.440** | -0.617** | 0.030 | -0.114 | 0.647** | 1 | ||
| LA | 0.005 | 0.085 | -0.110 | 0.122 | -0.132 | 0.022 | 0.081 | 1 | |
| LS | -0.082 | -0.093 | 0.022 | -0.093 | 0.182* | -0.089 | -0.155 | 0.033 | |
| SLB | 0.168 | 0.271** | -0.160 | 0.231* | -0.177* | 0.266** | 0.123 | 0.030 | |
| LBSUS | -0.197* | -0.363** | 0.256** | -0.191* | 0.197* | -0.378** | -0.163 | -0.161 | |
| LLB | 0.237** | 0.503** | -0.414** | 0.234** | -0.249** | 0.561** | 0.420** | 0.086 | |
| WLB | 0.154 | 0.411** | -0.397** | 0.185* | -0.149 | 0.458** | 0.358** | 0.018 | |
| LP | 0.357** | 0.493** | -0.240** | 0.388** | -0.263** | 0.469** | 0.189* | 0.034 | |
| LT | -0.034 | 0.030 | -0.078 | -0.014 | 0.083 | 0.069 | 0.118 | 0.022 | |
| DSP | 0 | 0.102 | -0.121 | 0.164 | -0.064 | 0.065 | 0 | -0.115 | |
| FCW | 0.246** | 0.467** | -0.330** | 0.377** | -0.183* | 0.442** | 0.163 | 0.037 | |
| 性状 Trait | 叶形 Leaf shape (LS) | 叶基形状Shape of leaf base(SLB) | 叶面形态Leaf blade:Shape of upper side (LBSUS) | 叶片长 Length of leaf blade(LLB) | 叶片宽 Width of leaf blade(WLB) | 叶柄长Length of petiole (LP) | 叶片厚 Leaf thickness (LT) | 花支轴紧密度Density of secondary peduncles(DSP) | 花序宽Flower cluster width(FCW) |
| LS | 1 | ||||||||
| SLB | -0.123 | 1 | |||||||
| LBSUS | -0.059 | -0.034 | 1 | ||||||
| LLB | 0.024 | 0.052 | -0.480** | 1 | |||||
| WLB | 0.269** | -0.009 | -0.355** | 0.866** | 1 | ||||
| LP | -0.054 | 0.03 | -0.139 | 0.538** | 0.456** | 1 | |||
| LT | -0.021 | -0.057 | -0.050 | 0.084 | 0.099 | -0.091 | 1 | ||
| DSP | 0.002 | 0.042 | 0.063 | 0.117 | 0.131 | 0.226* | 0.008 | 1 | |
| FCW | 0.057 | 0.146 | -0.375** | 0.579** | 0.549** | 0.433** | 0.096 | 0.544** | 1 |
Table 2 Correlation analysis of various traits in the F1 population of loquat
| 性状 Trait | 果纵径Longitudinal diameter (LD) | 果横径Transverse diameter (TD) | 果形指数Fruit shape index (FSI) | 果肉厚 Flesh thickness (FT) | 可溶性固形物Total soluble solids (TSS) | 种子质量 Seed weight (SW) | 种子数 Seed number (SN) | 叶姿 Leaf attitude(LA) | |
|---|---|---|---|---|---|---|---|---|---|
| LD | 1 | ||||||||
| TD | 0.735** | 1 | |||||||
| FSI | 0.248** | -0.472** | 1 | ||||||
| FT | 0.766** | 0.822** | -0.178* | 1 | |||||
| TSS | -0.440** | -0.496** | 0.136 | -0.493** | 1 | ||||
| SW | 0.568** | 0.894** | -0.532** | 0.517** | -0.322** | 1 | |||
| SN | 0.016 | 0.440** | -0.617** | 0.030 | -0.114 | 0.647** | 1 | ||
| LA | 0.005 | 0.085 | -0.110 | 0.122 | -0.132 | 0.022 | 0.081 | 1 | |
| LS | -0.082 | -0.093 | 0.022 | -0.093 | 0.182* | -0.089 | -0.155 | 0.033 | |
| SLB | 0.168 | 0.271** | -0.160 | 0.231* | -0.177* | 0.266** | 0.123 | 0.030 | |
| LBSUS | -0.197* | -0.363** | 0.256** | -0.191* | 0.197* | -0.378** | -0.163 | -0.161 | |
| LLB | 0.237** | 0.503** | -0.414** | 0.234** | -0.249** | 0.561** | 0.420** | 0.086 | |
| WLB | 0.154 | 0.411** | -0.397** | 0.185* | -0.149 | 0.458** | 0.358** | 0.018 | |
| LP | 0.357** | 0.493** | -0.240** | 0.388** | -0.263** | 0.469** | 0.189* | 0.034 | |
| LT | -0.034 | 0.030 | -0.078 | -0.014 | 0.083 | 0.069 | 0.118 | 0.022 | |
| DSP | 0 | 0.102 | -0.121 | 0.164 | -0.064 | 0.065 | 0 | -0.115 | |
| FCW | 0.246** | 0.467** | -0.330** | 0.377** | -0.183* | 0.442** | 0.163 | 0.037 | |
| 性状 Trait | 叶形 Leaf shape (LS) | 叶基形状Shape of leaf base(SLB) | 叶面形态Leaf blade:Shape of upper side (LBSUS) | 叶片长 Length of leaf blade(LLB) | 叶片宽 Width of leaf blade(WLB) | 叶柄长Length of petiole (LP) | 叶片厚 Leaf thickness (LT) | 花支轴紧密度Density of secondary peduncles(DSP) | 花序宽Flower cluster width(FCW) |
| LS | 1 | ||||||||
| SLB | -0.123 | 1 | |||||||
| LBSUS | -0.059 | -0.034 | 1 | ||||||
| LLB | 0.024 | 0.052 | -0.480** | 1 | |||||
| WLB | 0.269** | -0.009 | -0.355** | 0.866** | 1 | ||||
| LP | -0.054 | 0.03 | -0.139 | 0.538** | 0.456** | 1 | |||
| LT | -0.021 | -0.057 | -0.050 | 0.084 | 0.099 | -0.091 | 1 | ||
| DSP | 0.002 | 0.042 | 0.063 | 0.117 | 0.131 | 0.226* | 0.008 | 1 | |
| FCW | 0.057 | 0.146 | -0.375** | 0.579** | 0.549** | 0.433** | 0.096 | 0.544** | 1 |
| 性状 Trait | QTL数量 QTL number | 涉及连锁群 Involved LG | 贡献率/% Explain rate | LOD (0.995) |
|---|---|---|---|---|
| 果横径Transverse diameter | 26 | LG3、LG4、LG8、LG10、LG12、LG14、LG15、LG16、LG17 | 19.4 ~ 56.9 | 6.1 |
| 果纵径Longitudinal diameter | 8 | LG8、LG12、LG15、 | 18.3 ~ 22.2 | 5.7 |
| 果肉厚Flesh thickness | 15 | LG4、LG8、LG12、LG14、LG15、LG17 | 18.0 ~ 28.1 | 5.6 |
| 可溶性固形物Total soluble solids | 1 | LG12 | 19.4 ~ 20.3 | 6.0 |
| 种子质量Seed weight | 20 | LG3、LG4、LG8、LG10、LG12、LG14、LG15、LG16、LG17 | 17.7 ~ 61.5 | 5.5 |
| 种子数Seed number | 14 | LG4、LG8、LG12、LG16 | 18.7 ~ 25.5 | 5.8 |
| 果形指数Fruit shape index | 11 | LG3、LG4、LG8、LG12、LG14、LG15、LG16 | 18.1 ~ 35.8 | 5.6 |
| 花序支轴紧密度 Density of secondary peduncles | 2 | LG6、LG14 | 9.1 ~ 9.8 | 2.5 |
| 花序宽Flower cluster width | 7 | LG4、LG8、LG12、LG15 | 19.6 ~ 25.5 | 5.8 |
| 叶姿Leaf attitude | 2 | LG10、LG12 | 10.6 ~ 12.2 | 3.0 |
| 叶面形态 Leaf blade:Shape of upper side | 1 | LG12 | 22.9 ~ 23.9 | 6.4 |
| 叶形Leaf shape | 2 | LG4 | 10.0 ~ 11.9 | 3.0 |
| 叶片厚Leaf thickness | 1 | LG1 | 20.0 | 5.6 |
| 叶片长Length of leaf blade | 9 | LG3、LG4、LG8、LG12、LG15 | 19.9 ~ 37.5 | 5.9 |
| 叶柄长Length of petiole | 2 | LG12、LG16 | 19.7 ~ 22.2 | 5.8 |
| 叶基形状Shape of leaf base | 24 | LG3、LG4、LG8、LG10、LG12、LG15、LG16 | 19.2 ~ 41.7 | 5.7 |
| 叶片宽Width of leaf blade | 8 | LG8、LG12、LG15 | 21.2 ~ 30.7 | 6.3 |
Table 3 Summary of QTL mapping results
| 性状 Trait | QTL数量 QTL number | 涉及连锁群 Involved LG | 贡献率/% Explain rate | LOD (0.995) |
|---|---|---|---|---|
| 果横径Transverse diameter | 26 | LG3、LG4、LG8、LG10、LG12、LG14、LG15、LG16、LG17 | 19.4 ~ 56.9 | 6.1 |
| 果纵径Longitudinal diameter | 8 | LG8、LG12、LG15、 | 18.3 ~ 22.2 | 5.7 |
| 果肉厚Flesh thickness | 15 | LG4、LG8、LG12、LG14、LG15、LG17 | 18.0 ~ 28.1 | 5.6 |
| 可溶性固形物Total soluble solids | 1 | LG12 | 19.4 ~ 20.3 | 6.0 |
| 种子质量Seed weight | 20 | LG3、LG4、LG8、LG10、LG12、LG14、LG15、LG16、LG17 | 17.7 ~ 61.5 | 5.5 |
| 种子数Seed number | 14 | LG4、LG8、LG12、LG16 | 18.7 ~ 25.5 | 5.8 |
| 果形指数Fruit shape index | 11 | LG3、LG4、LG8、LG12、LG14、LG15、LG16 | 18.1 ~ 35.8 | 5.6 |
| 花序支轴紧密度 Density of secondary peduncles | 2 | LG6、LG14 | 9.1 ~ 9.8 | 2.5 |
| 花序宽Flower cluster width | 7 | LG4、LG8、LG12、LG15 | 19.6 ~ 25.5 | 5.8 |
| 叶姿Leaf attitude | 2 | LG10、LG12 | 10.6 ~ 12.2 | 3.0 |
| 叶面形态 Leaf blade:Shape of upper side | 1 | LG12 | 22.9 ~ 23.9 | 6.4 |
| 叶形Leaf shape | 2 | LG4 | 10.0 ~ 11.9 | 3.0 |
| 叶片厚Leaf thickness | 1 | LG1 | 20.0 | 5.6 |
| 叶片长Length of leaf blade | 9 | LG3、LG4、LG8、LG12、LG15 | 19.9 ~ 37.5 | 5.9 |
| 叶柄长Length of petiole | 2 | LG12、LG16 | 19.7 ~ 22.2 | 5.8 |
| 叶基形状Shape of leaf base | 24 | LG3、LG4、LG8、LG10、LG12、LG15、LG16 | 19.2 ~ 41.7 | 5.7 |
| 叶片宽Width of leaf blade | 8 | LG8、LG12、LG15 | 21.2 ~ 30.7 | 6.3 |
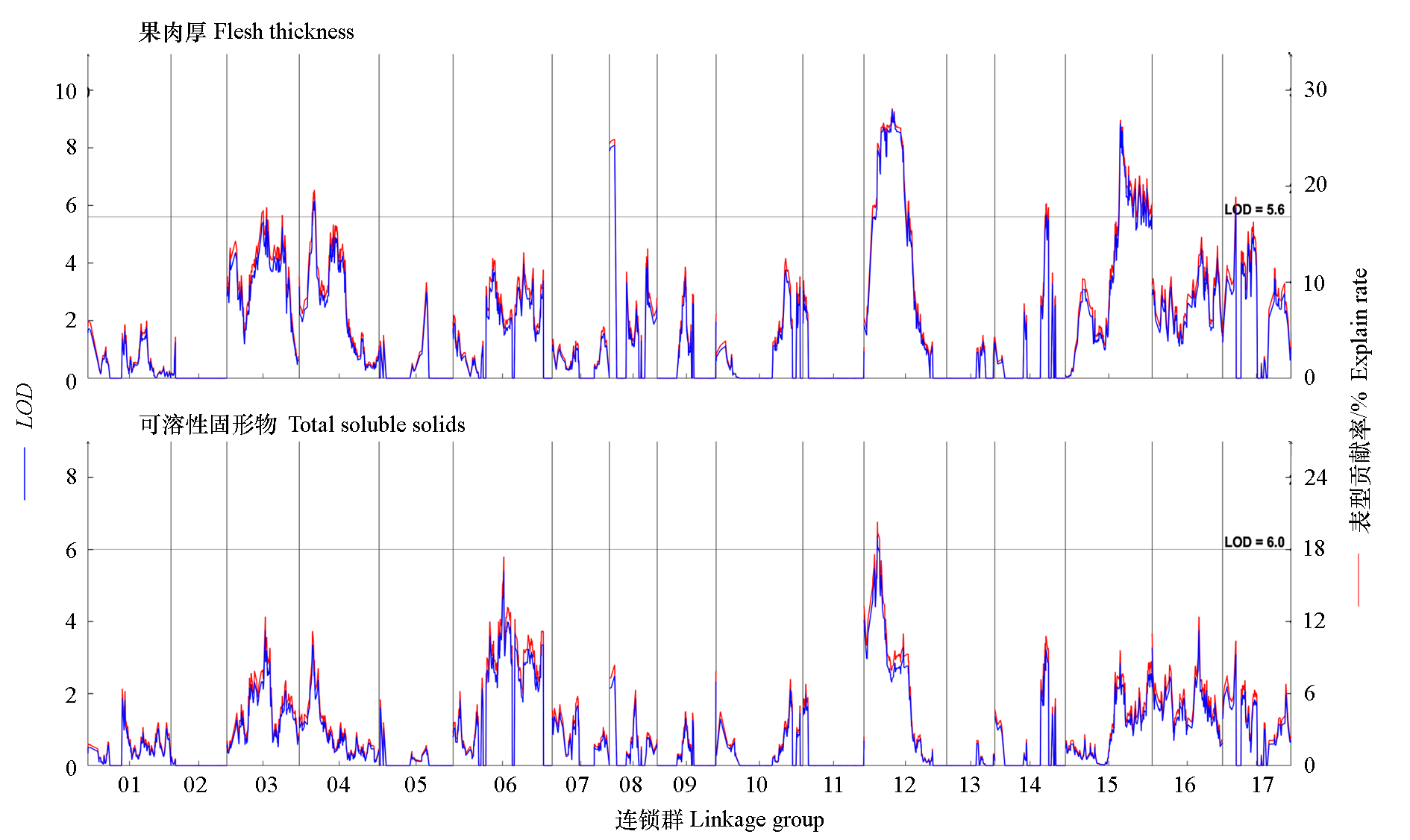
Fig. 2 The distribution of QTL for pulp thickness and total soluble solids on the linkage groups The horizontal grey line indicates the threshold of LOD with 0.995 confidence interval.
| 连锁群 LG | 重合区间/cM Overlapping intervals | 标记数量 Marker number | 性状 Trait | 对应的QTL Corresponding QTL |
|---|---|---|---|---|
| 4 | 13.500 ~ 14.276 | 2 | 花序宽、果形指数、叶片长、叶基形状、种子质量、果横径 Flower cluster width,fruit shape index,length of leaf blade,shape of leaf base,seed weight,transverse diameter | FCW.4.1、FSI.4.3、LLB.4.1、SLB.4.1、SW.4.1、TD.4.3 |
| 23.646 ~ 24.804 | 5 | 果形指数、果肉厚、叶片长、叶基形状、种子质量、果横径 Fruit shape index,flesh thickness,length of leaf blade,shape of leaf base,seed weight,transverse diameter | FSI.4.3、FT.4.1、LLB.4.1、SLB.4.1、SW.4.1、TD.4.3 | |
| 8 | 0 ~ 8.851 | 5 | 果形指数、果肉厚、果纵径、叶片长、叶基形状、种子质量、果横径 Fruit shape index,flesh thickness,longitudinal diameter,length of leaf blade,shape of leaf base,seed weight,transverse diameter | FSI.8.1、FT.8.1、LD.8.1、LLB.8.1、SLB.8.1、SW.8.1、TD.8.1 |
| 12 | 41.198 ~ 45.852 | 13 | 花序宽、果形指数、果肉厚、叶片长、叶柄长、叶基形状、种子数、种子质量、果横径、叶片宽 Flower cluster width,fruit shape index,flesh thickness,length of leaf blade,length of petiole,shape of leaf base,seed number,seed weight,transverse diameter,width of leaf blade | FCW.12.1、FSI.12.2、FT.12.3、LLB.12.1、LP.12.1、SLB.12.1、SN.12.1、SW.12.1、TD.12.1、WLB.12.1 |
| 65.246 ~ 66.407 | 7 | 果形指数、果肉厚、叶片长、叶基形状、种子数、种子质量、果横径、叶片宽 Fruit shape index,flesh thickness,length of leaf blade,shape of leaf base,seed number,seed weight,transverse diameter,width of leaf blade | FSI.12.2、FT.12.3、LLB.12.1、SLB.12.1、SN.12.1、SW.12.1、TD.12.1、WLB.12.3 | |
| 15 | 92.921 ~ 95.244 | 15 | 花序宽、果形指数、果肉厚、叶片长、叶基形状、种子质量、果横径、叶片宽 Flower cluster width,fruit shape index,flesh thickness,length of leaf blade,shape of leaf base,seed weight,transverse diameter,width of leaf blade | FCW15.2、FSI15.1、FT.15.1、LLB15.1、SLB.15.6、SW.15.2、TD.15.2、WLB15.2 |
Table 4 QTL cluster information
| 连锁群 LG | 重合区间/cM Overlapping intervals | 标记数量 Marker number | 性状 Trait | 对应的QTL Corresponding QTL |
|---|---|---|---|---|
| 4 | 13.500 ~ 14.276 | 2 | 花序宽、果形指数、叶片长、叶基形状、种子质量、果横径 Flower cluster width,fruit shape index,length of leaf blade,shape of leaf base,seed weight,transverse diameter | FCW.4.1、FSI.4.3、LLB.4.1、SLB.4.1、SW.4.1、TD.4.3 |
| 23.646 ~ 24.804 | 5 | 果形指数、果肉厚、叶片长、叶基形状、种子质量、果横径 Fruit shape index,flesh thickness,length of leaf blade,shape of leaf base,seed weight,transverse diameter | FSI.4.3、FT.4.1、LLB.4.1、SLB.4.1、SW.4.1、TD.4.3 | |
| 8 | 0 ~ 8.851 | 5 | 果形指数、果肉厚、果纵径、叶片长、叶基形状、种子质量、果横径 Fruit shape index,flesh thickness,longitudinal diameter,length of leaf blade,shape of leaf base,seed weight,transverse diameter | FSI.8.1、FT.8.1、LD.8.1、LLB.8.1、SLB.8.1、SW.8.1、TD.8.1 |
| 12 | 41.198 ~ 45.852 | 13 | 花序宽、果形指数、果肉厚、叶片长、叶柄长、叶基形状、种子数、种子质量、果横径、叶片宽 Flower cluster width,fruit shape index,flesh thickness,length of leaf blade,length of petiole,shape of leaf base,seed number,seed weight,transverse diameter,width of leaf blade | FCW.12.1、FSI.12.2、FT.12.3、LLB.12.1、LP.12.1、SLB.12.1、SN.12.1、SW.12.1、TD.12.1、WLB.12.1 |
| 65.246 ~ 66.407 | 7 | 果形指数、果肉厚、叶片长、叶基形状、种子数、种子质量、果横径、叶片宽 Fruit shape index,flesh thickness,length of leaf blade,shape of leaf base,seed number,seed weight,transverse diameter,width of leaf blade | FSI.12.2、FT.12.3、LLB.12.1、SLB.12.1、SN.12.1、SW.12.1、TD.12.1、WLB.12.3 | |
| 15 | 92.921 ~ 95.244 | 15 | 花序宽、果形指数、果肉厚、叶片长、叶基形状、种子质量、果横径、叶片宽 Flower cluster width,fruit shape index,flesh thickness,length of leaf blade,shape of leaf base,seed weight,transverse diameter,width of leaf blade | FCW15.2、FSI15.1、FT.15.1、LLB15.1、SLB.15.6、SW.15.2、TD.15.2、WLB15.2 |
| 性状 Trait | LG4: 13.500 ~ 14.276 cM | LG4: 23.646 ~ 24.804 cM | LG8: 0 ~ 8.851 cM | LG12: 41.198 ~ 45.852 cM | LG12: 65.246 ~ 66.407 cM | LG15: 92.921 ~ 95.244 cM |
|---|---|---|---|---|---|---|
| 叶基形状 Shape of leaf base | 26.1 ~ 27.4 | 22.3 ~ 23.3 | 38.7 ~ 41.7 | 35.9 ~ 37.1 | 27.7 ~ 28.8 | 31.5 ~ 34.2 |
| 种子质量 Seed weight | 30.4 ~ 31.4 | 37.6 ~ 39.9 | 54.7 ~ 56.0 | 58.0 ~ 58.8 | 42.8 ~ 45.3 | 45.4 ~ 47.0 |
| 果横径 Transverse diameter | 24.4 ~ 25.9 | 35.9 ~ 38.1 | 50.7 ~ 51.7 | 54.4 ~ 55.7 | 42.5 ~ 46.6 | 45.7 ~ 46.1 |
| 叶片长Length of leaf blade | 20.2 ~ 21.6 | 23.1 ~ 25.2 | 27.5 ~ 30.2 | 36.0 ~ 37.5 | 29.4 ~ 31.0 | 29.1 ~ 30.2 |
| 果形指数Fruit shape index | 18.5 ~ 20.5 | 23.9 ~ 24.7 | 22.4 ~ 24.4 | 30.2 ~ 30.7 | 25.1 ~ 30.3 | 24.3 ~ 25.0 |
| 果肉厚 Flesh thickness | 19.6 ~ 19.4 | 24.4 ~ 24.9 | 26.0 ~ 27.3 | 20.7 ~ 23.4 | 23.9 ~ 26.2 | |
| 叶片宽Width of leaf blade | 27.5 ~ 28.7 | 23.5 ~ 24.7 | 22.6 ~ 23.1 | |||
| 花序宽Flower cluster width | 19.9 ~ 22.0 | 22.2 ~ 23.9 | 20.3 ~ 21.5 | |||
| 种子数 Seed number | 21.5 ~ 22.3 | 21.6 ~ 24.5 | ||||
| 叶柄长Length of petiole | 19.7 ~ 22.2 | |||||
| 果纵径 Longitudinal diameter | 19.2 ~ 19.4 |
Table 5 Statistics of the explain rate of coincidence interval %
| 性状 Trait | LG4: 13.500 ~ 14.276 cM | LG4: 23.646 ~ 24.804 cM | LG8: 0 ~ 8.851 cM | LG12: 41.198 ~ 45.852 cM | LG12: 65.246 ~ 66.407 cM | LG15: 92.921 ~ 95.244 cM |
|---|---|---|---|---|---|---|
| 叶基形状 Shape of leaf base | 26.1 ~ 27.4 | 22.3 ~ 23.3 | 38.7 ~ 41.7 | 35.9 ~ 37.1 | 27.7 ~ 28.8 | 31.5 ~ 34.2 |
| 种子质量 Seed weight | 30.4 ~ 31.4 | 37.6 ~ 39.9 | 54.7 ~ 56.0 | 58.0 ~ 58.8 | 42.8 ~ 45.3 | 45.4 ~ 47.0 |
| 果横径 Transverse diameter | 24.4 ~ 25.9 | 35.9 ~ 38.1 | 50.7 ~ 51.7 | 54.4 ~ 55.7 | 42.5 ~ 46.6 | 45.7 ~ 46.1 |
| 叶片长Length of leaf blade | 20.2 ~ 21.6 | 23.1 ~ 25.2 | 27.5 ~ 30.2 | 36.0 ~ 37.5 | 29.4 ~ 31.0 | 29.1 ~ 30.2 |
| 果形指数Fruit shape index | 18.5 ~ 20.5 | 23.9 ~ 24.7 | 22.4 ~ 24.4 | 30.2 ~ 30.7 | 25.1 ~ 30.3 | 24.3 ~ 25.0 |
| 果肉厚 Flesh thickness | 19.6 ~ 19.4 | 24.4 ~ 24.9 | 26.0 ~ 27.3 | 20.7 ~ 23.4 | 23.9 ~ 26.2 | |
| 叶片宽Width of leaf blade | 27.5 ~ 28.7 | 23.5 ~ 24.7 | 22.6 ~ 23.1 | |||
| 花序宽Flower cluster width | 19.9 ~ 22.0 | 22.2 ~ 23.9 | 20.3 ~ 21.5 | |||
| 种子数 Seed number | 21.5 ~ 22.3 | 21.6 ~ 24.5 | ||||
| 叶柄长Length of petiole | 19.7 ~ 22.2 | |||||
| 果纵径 Longitudinal diameter | 19.2 ~ 19.4 |
| 连锁群 LG | 重合区间/cM Overlapping intervalsl | 基因数 Gene number | 注释Annotation | 总计Total | ||||
|---|---|---|---|---|---|---|---|---|
| COG | GO | KEGG | Swiss-Prot | Nr | ||||
| 4 | 13.500 ~ 14.276 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23.646 ~ 24.804 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 8 | 0 ~ 8.851 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 41.198 ~ 45.852 | 163 | 56 | 72 | 53 | 103 | 157 | 157 |
| 65.246 ~ 66.407 | 27 | 9 | 13 | 8 | 15 | 27 | 27 | |
| 15 | 92.921 ~ 95.244 | 24 | 3 | 5 | 7 | 12 | 24 | 24 |
Table 6 Statistics of gene annotations for coincidence interval
| 连锁群 LG | 重合区间/cM Overlapping intervalsl | 基因数 Gene number | 注释Annotation | 总计Total | ||||
|---|---|---|---|---|---|---|---|---|
| COG | GO | KEGG | Swiss-Prot | Nr | ||||
| 4 | 13.500 ~ 14.276 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23.646 ~ 24.804 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 8 | 0 ~ 8.851 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 41.198 ~ 45.852 | 163 | 56 | 72 | 53 | 103 | 157 | 157 |
| 65.246 ~ 66.407 | 27 | 9 | 13 | 8 | 15 | 27 | 27 | |
| 15 | 92.921 ~ 95.244 | 24 | 3 | 5 | 7 | 12 | 24 | 24 |
| QTL簇 QTL clusters | 区间/cM Interval | QTL | 候选基因ID Candidate gene ID | 功能注释 Function annotation |
|---|---|---|---|---|
| 12 | 41.198 ~ 45.852 | SW.12.1 SLB.12.1 TD.12.1 FT.12.3 LLB.12.1 FSI.12.2 WLB.12.1 FCW.12.1 SN.12.1 LP.12.1 | EVM0026248.1 | 含螺旋结构域蛋白SCD2 Coiled-coil domain-containing protein SCD2 |
| EVM0016634.2 | CRC结构域蛋白TSO1 CRC domain-containing protein TSO1 | |||
| EVM0021421.1 | F-box蛋白At1g55000 F-box protein At1g55000 | |||
| EVM0021006.1 | FRIGIDA-like蛋白1 FRIGIDA-like protein 1 | |||
| EVM0036521.1 | FRIGIDA-like蛋白3 FRIGIDA-like protein 3 | |||
| EVM0002631.1 | 赤霉素受体GID1C Gibberellin receptor GID1C | |||
| EVM0014419.1 | 谷胱甘肽合成酶,叶绿体(前体) Glutathione synthetase,chloroplastic(precursor) | |||
| EVM0007148.1 | 富甘氨酸RNA结合蛋白RZ1A Glycine-rich RNA-binding protein RZ1A | |||
| EVM0008020.1 | GTP结合蛋白At3g49725,叶绿体(前体) GTP-binding protein At3g49725,chloroplastic(precursor) | |||
| EVM0003720.1 | AUXIN RESPONSE 4蛋白 Protein AUXIN RESPONSE 4 | |||
| 15 | 92.921 ~ 95.244 | SW.15.2 TD.15.2 SLB.15.6 LLB.15.1 | EVM0039475.1 | 含硫氧还蛋白TTL1的TPR重复序列 TPR repeat-containing thioredoxin TTL1 |
| EVM0017898.1 | 含有甲基CpG结合结构域蛋白9 | |||
| Methyl-CpG-binding domain-containing protein 9 | ||||
| FT.15.1 FSI.15.1 WLB.15.2 FCW.15.2 | EVM0022488.1 | Knotted-1-like 同源盒蛋白3 Homeobox protein knotted-1-like 3 |
Table 7 QTL clusters and candidate gene functional predictions
| QTL簇 QTL clusters | 区间/cM Interval | QTL | 候选基因ID Candidate gene ID | 功能注释 Function annotation |
|---|---|---|---|---|
| 12 | 41.198 ~ 45.852 | SW.12.1 SLB.12.1 TD.12.1 FT.12.3 LLB.12.1 FSI.12.2 WLB.12.1 FCW.12.1 SN.12.1 LP.12.1 | EVM0026248.1 | 含螺旋结构域蛋白SCD2 Coiled-coil domain-containing protein SCD2 |
| EVM0016634.2 | CRC结构域蛋白TSO1 CRC domain-containing protein TSO1 | |||
| EVM0021421.1 | F-box蛋白At1g55000 F-box protein At1g55000 | |||
| EVM0021006.1 | FRIGIDA-like蛋白1 FRIGIDA-like protein 1 | |||
| EVM0036521.1 | FRIGIDA-like蛋白3 FRIGIDA-like protein 3 | |||
| EVM0002631.1 | 赤霉素受体GID1C Gibberellin receptor GID1C | |||
| EVM0014419.1 | 谷胱甘肽合成酶,叶绿体(前体) Glutathione synthetase,chloroplastic(precursor) | |||
| EVM0007148.1 | 富甘氨酸RNA结合蛋白RZ1A Glycine-rich RNA-binding protein RZ1A | |||
| EVM0008020.1 | GTP结合蛋白At3g49725,叶绿体(前体) GTP-binding protein At3g49725,chloroplastic(precursor) | |||
| EVM0003720.1 | AUXIN RESPONSE 4蛋白 Protein AUXIN RESPONSE 4 | |||
| 15 | 92.921 ~ 95.244 | SW.15.2 TD.15.2 SLB.15.6 LLB.15.1 | EVM0039475.1 | 含硫氧还蛋白TTL1的TPR重复序列 TPR repeat-containing thioredoxin TTL1 |
| EVM0017898.1 | 含有甲基CpG结合结构域蛋白9 | |||
| Methyl-CpG-binding domain-containing protein 9 | ||||
| FT.15.1 FSI.15.1 WLB.15.2 FCW.15.2 | EVM0022488.1 | Knotted-1-like 同源盒蛋白3 Homeobox protein knotted-1-like 3 |
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
|
陈静, 王丹丹, 周芳玲, 彭泽, 杨向晖. 2023. 枇杷花序支轴紧密度量化指标初步研究及新型判别技术开发. 果树学报, 40 (7):1471-1485.
|
|
| [6] |
|
| [7] |
|
| [8] |
doi: 10.1126/science.1122847 pmid: 16690816 |
| [9] |
|
| [10] |
|
| [11] |
|
|
郭乙含, 赵崇斌, 李舒庆, 徐红霞, 黄天启, 林顺权, 陈俊伟, 杨向晖. 2023. 枇杷叶片性状与单果质量的遗传规律研究. 热带亚热带植物学报, 31 (1):62-68.
|
|
| [12] |
doi: 10.1242/dev.127.10.2219 pmid: 10769245 |
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
pmid: 8903506 |
| [19] |
|
|
林顺权. 2019. 两组词汇“枇杷”与“琵琶”、“枇杷”与“卢橘”史料考析. 果树学报, 36 (7):922-927.
|
|
| [20] |
doi: 10.16420/j.issn.0513-353x.2021-0248 |
|
吕正鑫, 贺艳群, 贾东峰, 黄春辉, 钟敏, 廖光联, 朱壹, 袁开昌, 刘传浩, 徐小彪. 2022. 猕猴桃种质资源表型性状遗传多样性分析. 园艺学报, 49 (7):1571-1581.
doi: 10.16420/j.issn.0513-353x.2021-0248 |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
doi: 10.1111/nph.16403 pmid: 31880321 |
| [27] |
doi: 10.16420/j.issn.0513-353x.2022-0056 |
|
唐海霞, 裴广营, 张琼, 王中堂. 2023. 枣果实相关性状QTL定位分析. 园艺学报, 50 (4):754-764.
doi: 10.16420/j.issn.0513-353x.2022-0056 |
|
| [28] |
doi: 10.1104/pp.18.01434 pmid: 30659066 |
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
doi: 10.1104/pp.15.00801 pmid: 26392261 |
| [33] |
|
|
许莉斯. 2012. 枣果实性状QTL定位及优良基因型筛选研究[硕士论文]. 保定: 河北农业大学.
|
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
|
张学超, 任海龙, 唐式敏, 朱玲, 张胜军, 冉昪. 2021. 伊犁天山160份野苹果种质资源表型性状的遗传多样性分析. 植物遗传资源学报, 22 (6):1521-1530.
doi: 10.13430/j.cnki.jpgr. 20210415002 |
|
| [38] |
|
|
赵崇斌, 郭乙含, 李舒庆, 徐红霞, 黄天启, 林顺权, 陈俊伟, 杨向晖. 2021. ‘宁海白’ב大房’枇杷F1杂交群体果实性状的相关性及遗传分析. 果树学报, 38 (7):1055-1065.
|
|
| [39] |
|
|
赵旗峰, 马小河, 董志刚, 唐晓萍, 陈俊. 2013. ‘秋红宝’葡萄植物学特征的杂种优势分析. 天津农业科学, 19 (6):84-86.
|
|
| [40] |
|
|
郑少泉, 陈秀萍, 许秀淡. 2006. 枇杷种质资源描述规范和数据标准. 北京: 中国农业出版社.
|
| [1] | XIA Zhilei, YU Bingxin, YANG Meng, LIAN Zilin, GUAN Lilan, HE Yichao, YAN Shuangshuang, CAO Bihao, and QIU Zhengkun, . Fine-Mapping and KASP Marker Development for the Gene Underlying Green Fruit in Eggplant [J]. Acta Horticulturae Sinica, 2024, 51(9): 2008-2018. |
| [2] | ZHAO Jiaying, ZENG Zhouting, CEN Xinying, SHI Jiaoqi, LI Xiaoxian, SHEN Xiaoxia, and YU Zhenming, . Genome-Wide Identification and Expression Analysis of CCO Gene Family in Dendrobium officinale During Flower Development [J]. Acta Horticulturae Sinica, 2024, 51(9): 2075-2088. |
| [3] | ZHONG Yizhong, WANG Zhou, XIE Jingyao, WANG Zhengpeng, HAO Jingjing, LIU Chaoyang, ZHANG Wei, WU Jing, ZHONG Ziqin, CHEN Chengjie, and HE Yehua, . Flowering and Fruiting Behaviour of Two South China Plum Cultivars with Different Ripening Stages in an Area Near the Northern Boundary of the Tropics [J]. Acta Horticulturae Sinica, 2024, 51(9): 2105-2119. |
| [4] | LIU Yanfei, HE Xin, TIAN Ailin, and LIU Zhande. A New High-Quality Kiwifruit Cultivar with Better Storability‘Jinfu’ [J]. Acta Horticulturae Sinica, 2024, 51(9): 2219-2220. |
| [5] | QIU Hui, ZHU Dejuan, ZHANG Yongle, GAO Yujie, LI Liu, WANG Guoping, HONG Ni. Interaction Between the Coat Protein of Apple Chlorotic Leaf Spot Virus and Two E3 Ubiquitin Ligases of Pear and Their Subcellular Localization [J]. Acta Horticulturae Sinica, 2024, 51(8): 1715-1727. |
| [6] | ZHAO Jingyi, WU Xiaoxu, HU Yunjie, GAI Shuting, ZHU Zhihao, QIN Lei, WANG Yong. Molecular Cloning and Functional Analysis of AcGAI in Onion Flowering Regulation [J]. Acta Horticulturae Sinica, 2024, 51(8): 1792-1802. |
| [7] | LI Xujiao, LÜ Qi, YAO Dongdong, ZHAO Fengyun, WANG Xiaofei, YU Kun. Effect of‘Yanfu 3’Apple Grafted with Different Rootstocks on Absorption and Utilization of 15N-urea [J]. Acta Horticulturae Sinica, 2024, 51(8): 1868-1880. |
| [8] | GUO Qunyan, LIU Caixian, YU Qiuxiu, ZHANG Hua, ZHENG Meiting. Studies on the Physiological Indexes and Endogenous Hormones Variation During Flower Development in Michelia crassipes [J]. Acta Horticulturae Sinica, 2024, 51(8): 1881-1890. |
| [9] | WEN Songqin, LI Jialin, CHI Zhuoheng, XIA Yan, WANG Shuming, WU Di, HAO Yawen, GUO Qigao, LIANG Guolu, JING Danlong. The Role of Light and Temperature Related Genes Alternative Splicing in Regulating Plant Flowering Time [J]. Acta Horticulturae Sinica, 2024, 51(8): 1949-1963. |
| [10] | ZHANG Lihua, XU Yu, Zheng Litong, Wang Changzhi, ZHU Lingcheng, MA Baiquan, and LI Mingjun, . The Relevance Research Between Acid Transporters and Fruit Acidity [J]. Acta Horticulturae Sinica, 2024, 51(7): 1474-1488. |
| [11] | LI Xin, CHAI Yingfang, TIAN Zhen, WANG Pengwei, and CHENG Yunjiang. Isolation and Purification of Mitochondria and Its Application in Research on Fruit Ripening and Senescence [J]. Acta Horticulturae Sinica, 2024, 51(7): 1516-1528. |
| [12] | HUANG Yiqi, ZHU Yuqin, DU Zhaoxuan, XU Feng, CHEN Xiaoyi, YANG Guoshun, and XU Yanshuai, . Progress on Bud Sport of Tree Fruit [J]. Acta Horticulturae Sinica, 2024, 51(7): 1547-1564. |
| [13] | LI Zhenjian, ZHANG Suzhi, ZOU Xiaoshuo, CHANG Lingling, ZHOU Yuxin, SUN Zhenyuan, and ZOU Junzhu, . A New Lycaste Cultivar‘Hong Linwu’ [J]. Acta Horticulturae Sinica, 2024, 51(7): 1713-1714. |
| [14] | ZHU Qixuan, LI Xiaoying, WU Junkai, GE Hang, CHEN Junwei, XU Hongxia. Genetic Tendency Analysis and Comprehensive Evaluation of the Fruit Traits in Loquat F1 Generation [J]. Acta Horticulturae Sinica, 2024, 51(6): 1201-1215. |
| [15] | SUN Zhichao, GUO Xinmiao, WANG Hui, XIE Yan, GAO Yanxia, LI Jisheng, GAO Yujun. Mechanism of miR408-MnBBP Module Regulating Anthocyanin Biosynthesis in Mulberry [J]. Acta Horticulturae Sinica, 2024, 51(6): 1216-1226. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd