
Acta Horticulturae Sinica ›› 2023, Vol. 50 ›› Issue (12): 2735-2747.doi: 10.16420/j.issn.0513-353x.2023-0277
• Plant Protection • Previous Articles Next Articles
CHEN Ling1, GUO Cheng1, JIA Anning2, DENG Congliang3, CHONG Yan3, SHI Xiju3, LI Yongqiang2,*( )
)
Received:2023-06-19
Revised:2023-10-24
Online:2023-12-25
Published:2023-12-29
Contact:
LI Yongqiang
CHEN Ling, GUO Cheng, JIA Anning, DENG Congliang, CHONG Yan, SHI Xiju, LI Yongqiang. Determination and Analyses of the Complete Genome Sequences of Apple Stem Grooving Virus Isolates Infecting Tree Peony[J]. Acta Horticulturae Sinica, 2023, 50(12): 2735-2747.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0277
| 产物/bp Product | 引物位置/nt Location | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence |
|---|---|---|---|
| 1 953 | 494 ~ 513 | ASGV-F1 | TGGTCTGAGGAACCAATGCC |
| 2 446 ~ 2 427 | ASGV-R1 | ATCCTCTTGACGTTTGGCGT | |
| 2 342 | 1 533 ~ 1 552 | ASGV-F2 | TCAAAGCTGGTCAGAGCGAG |
| 3 874 ~ 3 855 | ASGV-R2 | AAAGCATGGTCGAGCTTCCA | |
| 2 083 | 3 498 ~ 3 517 | ASGV-F3 | TCCATACTCGGCCTCTCCAA |
| 5 580 ~ 5 561 | ASGV-R3 | TTGGAGTTTGGCCCCCTTTT | |
| 1 413 ~ 1 392 | 5ʹ RACE | GATTACGCCAAGCTT-AACCTGCGGCCAAGCAGGTCCAA | |
| 5 128 ~ 5 152 | 3ʹ RACE | GATTACGCCAAGCTT-TGCCTGTTTGGACCCAGAAAGGGGT |
Table 1 The sequences of primers used
| 产物/bp Product | 引物位置/nt Location | 引物名称 Primer name | 引物序列(5′-3′) Primer sequence |
|---|---|---|---|
| 1 953 | 494 ~ 513 | ASGV-F1 | TGGTCTGAGGAACCAATGCC |
| 2 446 ~ 2 427 | ASGV-R1 | ATCCTCTTGACGTTTGGCGT | |
| 2 342 | 1 533 ~ 1 552 | ASGV-F2 | TCAAAGCTGGTCAGAGCGAG |
| 3 874 ~ 3 855 | ASGV-R2 | AAAGCATGGTCGAGCTTCCA | |
| 2 083 | 3 498 ~ 3 517 | ASGV-F3 | TCCATACTCGGCCTCTCCAA |
| 5 580 ~ 5 561 | ASGV-R3 | TTGGAGTTTGGCCCCCTTTT | |
| 1 413 ~ 1 392 | 5ʹ RACE | GATTACGCCAAGCTT-AACCTGCGGCCAAGCAGGTCCAA | |
| 5 128 ~ 5 152 | 3ʹ RACE | GATTACGCCAAGCTT-TGCCTGTTTGGACCCAGAAAGGGGT |
| 取样位置 Sampling location | 植株症状 Plant symptom | 分离物 Isolate | 基因组大小/nt Genome size | 登录号 Accession No. |
|---|---|---|---|---|
| 海淀公园Haidian Park | 褪绿Chlorosis | Peony BJ2 | 6 470 | OQ658829 |
| 卷叶Leafroll | Peony BJ3 | 6 470 | OQ658830 | |
| 景山公园Jingshan Park | 褪绿Chlorosis | Peony BJ4 | 6 469 | OQ658831 |
| 皱缩、节间缩短Crinkle,internode shortening | Peony BJ5 | 6 467 | OQ658832 | |
| 卷叶Leafroll | Peony BJ6 | 6 467 | OQ658833 | |
| 北京植物园Beijing Botanical Garden | 红叶Leaf reddening | Peony BJ7 | 6 495 | OQ658834 |
| 卷叶Leafroll | Peony BJ8 | 6 495 | OQ658835 | |
| 圆明园Yuanmingyuan Garden | 褪绿Chlorosis | Peony BJ9 | 6 469 | OQ658836 |
| 皱缩Crinkle | Peony BJ10 | 6 469 | OQ658837 |
Table 2 The information of the genome sequences of ASGV-tree peony isolates
| 取样位置 Sampling location | 植株症状 Plant symptom | 分离物 Isolate | 基因组大小/nt Genome size | 登录号 Accession No. |
|---|---|---|---|---|
| 海淀公园Haidian Park | 褪绿Chlorosis | Peony BJ2 | 6 470 | OQ658829 |
| 卷叶Leafroll | Peony BJ3 | 6 470 | OQ658830 | |
| 景山公园Jingshan Park | 褪绿Chlorosis | Peony BJ4 | 6 469 | OQ658831 |
| 皱缩、节间缩短Crinkle,internode shortening | Peony BJ5 | 6 467 | OQ658832 | |
| 卷叶Leafroll | Peony BJ6 | 6 467 | OQ658833 | |
| 北京植物园Beijing Botanical Garden | 红叶Leaf reddening | Peony BJ7 | 6 495 | OQ658834 |
| 卷叶Leafroll | Peony BJ8 | 6 495 | OQ658835 | |
| 圆明园Yuanmingyuan Garden | 褪绿Chlorosis | Peony BJ9 | 6 469 | OQ658836 |
| 皱缩Crinkle | Peony BJ10 | 6 469 | OQ658837 |
| 分离物Isolate | Peony BJ2 | Peony BJ3 | Peony BJ4 | Peony BJ5 | Peony BJ6 | Peony BJ | Peony BJ7 | Peony BJ8 | Peony BJ9 |
|---|---|---|---|---|---|---|---|---|---|
| Peony BJ3 | 96.4 | ||||||||
| Peony BJ4 | 87.4 | 87.6 | |||||||
| Peony BJ5 | 87.4 | 86.8 | 92.2 | ||||||
| Peony BJ6 | 83.3 | 83.5 | 90.3 | 97.0 | |||||
| Peony BJ | 82.1 | 82.1 | 90.2 | 87.3 | 88.7 | ||||
| Peony BJ7 | 83.2 | 83.1 | 83.0 | 87.4 | 86.5 | 87.3 | |||
| Peony BJ8 | 82.3 | 82.2 | 83.6 | 85.5 | 86.8 | 88.6 | 91.6 | ||
| Peony BJ9 | 94.8 | 95.2 | 87.0 | 86.4 | 83.5 | 81.9 | 83.1 | 82.2 | |
| Peony BJ10 | 94.8 | 95.1 | 86.8 | 86.1 | 83.2 | 82.0 | 83.0 | 82.2 | 97.6 |
| 其他Other | 82.4 ~ 912 | 82.3 ~ 91.3 | 81.9 ~ 86.8 | 81.8 ~ 87.2 | 82.6 ~ 87.9 | 81.2 ~ 89.0 | 82.1 ~ 90.5 | 80.8 ~ 91.0 | 82.0 ~ 90.7 |
Table 3 Pairwise alignment of genomic nucleotide sequences of ASGV-tree peony isolates and the identity of each genomic nucleotide sequence of ASGV-tree peony isolates with that of ASGV isolates from other plant species %
| 分离物Isolate | Peony BJ2 | Peony BJ3 | Peony BJ4 | Peony BJ5 | Peony BJ6 | Peony BJ | Peony BJ7 | Peony BJ8 | Peony BJ9 |
|---|---|---|---|---|---|---|---|---|---|
| Peony BJ3 | 96.4 | ||||||||
| Peony BJ4 | 87.4 | 87.6 | |||||||
| Peony BJ5 | 87.4 | 86.8 | 92.2 | ||||||
| Peony BJ6 | 83.3 | 83.5 | 90.3 | 97.0 | |||||
| Peony BJ | 82.1 | 82.1 | 90.2 | 87.3 | 88.7 | ||||
| Peony BJ7 | 83.2 | 83.1 | 83.0 | 87.4 | 86.5 | 87.3 | |||
| Peony BJ8 | 82.3 | 82.2 | 83.6 | 85.5 | 86.8 | 88.6 | 91.6 | ||
| Peony BJ9 | 94.8 | 95.2 | 87.0 | 86.4 | 83.5 | 81.9 | 83.1 | 82.2 | |
| Peony BJ10 | 94.8 | 95.1 | 86.8 | 86.1 | 83.2 | 82.0 | 83.0 | 82.2 | 97.6 |
| 其他Other | 82.4 ~ 912 | 82.3 ~ 91.3 | 81.9 ~ 86.8 | 81.8 ~ 87.2 | 82.6 ~ 87.9 | 81.2 ~ 89.0 | 82.1 ~ 90.5 | 80.8 ~ 91.0 | 82.0 ~ 90.7 |
| 分离物Isolate | 核苷酸序列Nucleotide sequence | 氨基酸序列Amino acid sequence | ||||||
|---|---|---|---|---|---|---|---|---|
| 5ʹ UTR | ORF1 | CP | ORF2 | 3ʹ UTR | Polyprotein | CP | MP | |
| Peony BJ2 | — | 81.8 ~ 91.1 | 92.9 ~ 97.8 | 85.8 ~ 92.9 | 96.5 ~ 100 | 84.1 ~ 94.5 | 96.2 ~ 98.7 | 92.5 ~ 98.4 |
| Peony BJ3 | — | 82.8 ~ 91.4 | 92.6 ~ 97.2 | 85.8 ~ 93.3 | 96.5 ~ 100 | 85.8 ~ 94.8 | 96.6 ~ 99.2 | 91.9 ~ 97.5 |
| Peony BJ4 | 100 | 81.5 ~ 86.6 | 91.2 ~ 94.0 | 85.3 ~ 88.4 | 95.6 ~ 99.1 | 86.7 ~ 92.1 | 94.1 ~ 97.9 | 92.8 ~ 98.1 |
| Peony BJ5 | 97.0 ~ 100 | 81.5 ~ 86.4 | 91.5 ~ 94.3 | 83.8 ~ 87.5 | 95.6 ~ 99.1 | 86.4 ~ 91.2 | 96.2 ~ 99.2 | 89.4 ~ 94.7 |
| Peony BJ6 | 97.1 ~ 100 | 82.3 ~ 87.7 | 91.9 ~ 94.1 | 85.3 ~ 88.7 | 95.6 ~ 99.1 | 86.5 ~ 90.9 | 94.9 ~ 98.3 | 92.2 ~ 98.1 |
| Peony BJ | 97.1 ~ 100 | 81.1 ~ 88.9 | 91.4 ~ 94.3 | 85.0 ~ 88.7 | 93.0 ~ 96.5 | 86.6 ~ 92.5 | 94.9 ~ 98.3 | 91.9 ~ 97.5 |
| Peony BJ7 | 97.1 ~ 100 | 80.6 ~ 90.4 | 91.5 ~ 93.6 | 85.5 ~ 92.8 | 95.1 ~ 99.3 | 85.6 ~ 91.9 | 94.1 ~ 97.1 | 92.5 ~ 98.1 |
| Peony BJ8 | 97.1 ~ 100 | 81.5 ~ 90.9 | 91.9 ~ 97.2 | 84.8 ~ 88.0 | 95.7 ~ 99.3 | 85.9 ~ 90.9 | 94.5 ~ 98.3 | 92.2 ~ 97.2 |
| Peony BJ9 | 97.1 ~ 100 | 81.7 ~ 90.6 | 91.7 ~ 98.6 | 86.0 ~ 92.9 | 94.8 ~ 99.1 | 85.7 ~ 94.0 | 94.5 ~ 98.7 | 92.8 ~ 98.8 |
| Peony BJ10 | 97.1 ~ 100 | 81.7 ~ 90.7 | 91.3 ~ 98.0 | 85.6 ~ 92.9 | 94.8 ~ 99.1 | 85.7 ~ 94.2 | 94.5 ~ 98.7 | 91.9 ~ 97.8 |
Table 4 The identity in nucleotides and their encoding proteins of ASGV-tree peony isolates with those of ASGV isolates from other plant species %
| 分离物Isolate | 核苷酸序列Nucleotide sequence | 氨基酸序列Amino acid sequence | ||||||
|---|---|---|---|---|---|---|---|---|
| 5ʹ UTR | ORF1 | CP | ORF2 | 3ʹ UTR | Polyprotein | CP | MP | |
| Peony BJ2 | — | 81.8 ~ 91.1 | 92.9 ~ 97.8 | 85.8 ~ 92.9 | 96.5 ~ 100 | 84.1 ~ 94.5 | 96.2 ~ 98.7 | 92.5 ~ 98.4 |
| Peony BJ3 | — | 82.8 ~ 91.4 | 92.6 ~ 97.2 | 85.8 ~ 93.3 | 96.5 ~ 100 | 85.8 ~ 94.8 | 96.6 ~ 99.2 | 91.9 ~ 97.5 |
| Peony BJ4 | 100 | 81.5 ~ 86.6 | 91.2 ~ 94.0 | 85.3 ~ 88.4 | 95.6 ~ 99.1 | 86.7 ~ 92.1 | 94.1 ~ 97.9 | 92.8 ~ 98.1 |
| Peony BJ5 | 97.0 ~ 100 | 81.5 ~ 86.4 | 91.5 ~ 94.3 | 83.8 ~ 87.5 | 95.6 ~ 99.1 | 86.4 ~ 91.2 | 96.2 ~ 99.2 | 89.4 ~ 94.7 |
| Peony BJ6 | 97.1 ~ 100 | 82.3 ~ 87.7 | 91.9 ~ 94.1 | 85.3 ~ 88.7 | 95.6 ~ 99.1 | 86.5 ~ 90.9 | 94.9 ~ 98.3 | 92.2 ~ 98.1 |
| Peony BJ | 97.1 ~ 100 | 81.1 ~ 88.9 | 91.4 ~ 94.3 | 85.0 ~ 88.7 | 93.0 ~ 96.5 | 86.6 ~ 92.5 | 94.9 ~ 98.3 | 91.9 ~ 97.5 |
| Peony BJ7 | 97.1 ~ 100 | 80.6 ~ 90.4 | 91.5 ~ 93.6 | 85.5 ~ 92.8 | 95.1 ~ 99.3 | 85.6 ~ 91.9 | 94.1 ~ 97.1 | 92.5 ~ 98.1 |
| Peony BJ8 | 97.1 ~ 100 | 81.5 ~ 90.9 | 91.9 ~ 97.2 | 84.8 ~ 88.0 | 95.7 ~ 99.3 | 85.9 ~ 90.9 | 94.5 ~ 98.3 | 92.2 ~ 97.2 |
| Peony BJ9 | 97.1 ~ 100 | 81.7 ~ 90.6 | 91.7 ~ 98.6 | 86.0 ~ 92.9 | 94.8 ~ 99.1 | 85.7 ~ 94.0 | 94.5 ~ 98.7 | 92.8 ~ 98.8 |
| Peony BJ10 | 97.1 ~ 100 | 81.7 ~ 90.7 | 91.3 ~ 98.0 | 85.6 ~ 92.9 | 94.8 ~ 99.1 | 85.7 ~ 94.2 | 94.5 ~ 98.7 | 91.9 ~ 97.8 |

Fig. 1 Schematic representation of the genome structure of ASGV-tree peony isolate(Peony BJ5) The numbers in the figure represent the nucleotide location of ORF1,ORF2 and each conserved domain in the polyprotein encoded by ORF1.

Fig. 2 The complete genome sequence alignment of ASGV-tree peony isolates(MT607622,OQ658829-OQ658837)with ASGV other host plants isolates MK599422(loquat,China),LC475148(pear,South Korea),LC480457(Cnidium officinale,South Korea),MZ330115(citrus,USA),OP535347(apple,Brazil).
| 重组序列 Recombination sequence | 主、次要亲本 Major and minor parents | 重组发生的比对起、 终点/nt Breakpoint positions in alignment(Begin,End) | RDP v.4.101软件中7种检测方法的P值 | |||
|---|---|---|---|---|---|---|
| P-value for the seven detection methods in RDP v.4.101 | ||||||
| RDP GENECONV | BootScan MaxChi | Chimaera SiScan | 3Seq | |||
| OQ658832 | OQ658833 OQ658829 | 待定Undetermined 2 022 | 3.064 × 10-87 3.681 × 10-104 | 3.083 × 10-100 1.879 × 10-42 | 3.451 × 10-45 5.539 × 10-46 | 2.963 × 10-11 |
| OQ658831 | MT607622 OQ658829 | 待定Undetermined 1 998 | 1.256 × 10-58 9.314 × 10-73 | 3.861 × 10-64 1.142 × 10-32 | 6.943 × 10-37 2.647 × 10-39 | 5.926 × 10-11 |
| OQ658833 | 未知Unknown MT607622 | 4 084 待定Undetermined | 1.174 × 10-41 2.230 × 10-36 | 3.444 × 10-21 1.725 × 10-18 | 5.191 × 10-14 4.712 × 10-32 | 2.963 × 10-11 |
| OQ658831 | MT607622 未知Unknown | 待定Undetermined 2 446 | 1.221 × 10-30 3.455 × 10-17 | 9.406 × 10-27 9.470 × 10-11 | 5.297 × 10-12 2.231 × 10-9 | 5.926 × 10-11 |
| MT607622 | 未知Unknown OQ658835 | 待定Undetermined 2 550 | 1.716 × 10-13 8.543 × 10-6 | 1.459 × 10-12 2.437 × 10-15 | 9.076 × 10-12 3.670 × 10-29 | 6.118 × 10-23 |
| OQ658835 | OQ658834 MK929792 | 5 532 待定Undetermined | 1.740 × 10-22 1.291 × 10-12 | 1.736 × 10-22 — | 1.244 × 10-6 4.068 × 10-11 | 5.926 × 10-11 |
Table 5 The information of the recombination events of ASGV-tree peony isolates
| 重组序列 Recombination sequence | 主、次要亲本 Major and minor parents | 重组发生的比对起、 终点/nt Breakpoint positions in alignment(Begin,End) | RDP v.4.101软件中7种检测方法的P值 | |||
|---|---|---|---|---|---|---|
| P-value for the seven detection methods in RDP v.4.101 | ||||||
| RDP GENECONV | BootScan MaxChi | Chimaera SiScan | 3Seq | |||
| OQ658832 | OQ658833 OQ658829 | 待定Undetermined 2 022 | 3.064 × 10-87 3.681 × 10-104 | 3.083 × 10-100 1.879 × 10-42 | 3.451 × 10-45 5.539 × 10-46 | 2.963 × 10-11 |
| OQ658831 | MT607622 OQ658829 | 待定Undetermined 1 998 | 1.256 × 10-58 9.314 × 10-73 | 3.861 × 10-64 1.142 × 10-32 | 6.943 × 10-37 2.647 × 10-39 | 5.926 × 10-11 |
| OQ658833 | 未知Unknown MT607622 | 4 084 待定Undetermined | 1.174 × 10-41 2.230 × 10-36 | 3.444 × 10-21 1.725 × 10-18 | 5.191 × 10-14 4.712 × 10-32 | 2.963 × 10-11 |
| OQ658831 | MT607622 未知Unknown | 待定Undetermined 2 446 | 1.221 × 10-30 3.455 × 10-17 | 9.406 × 10-27 9.470 × 10-11 | 5.297 × 10-12 2.231 × 10-9 | 5.926 × 10-11 |
| MT607622 | 未知Unknown OQ658835 | 待定Undetermined 2 550 | 1.716 × 10-13 8.543 × 10-6 | 1.459 × 10-12 2.437 × 10-15 | 9.076 × 10-12 3.670 × 10-29 | 6.118 × 10-23 |
| OQ658835 | OQ658834 MK929792 | 5 532 待定Undetermined | 1.740 × 10-22 1.291 × 10-12 | 1.736 × 10-22 — | 1.244 × 10-6 4.068 × 10-11 | 5.926 × 10-11 |
| 序列 Sequence | 分离物 Isolate | 寄主 Host | 地理位置 Geographical region |
|---|---|---|---|
| OQ658829 | Peony BJ2 | 牡丹Tree peony | 中国China |
| OQ658831 | Peony BJ4 | 牡丹Tree peony | 中国China |
| OQ658832 | Peony BJ5 | 牡丹Tree peony | 中国China |
| OQ658833 | Peony BJ6 | 牡丹Tree peony | 中国China |
| OQ658834 | Peony BJ7 | 牡丹Tree peony | 中国China |
| OQ658835 | Peony BJ8 | 牡丹Tree peony | 中国China |
| MT607622 | Peony BJ | 牡丹Tree peony | 中国China |
| MK929792 | BR-Brael | 苹果Apple | 巴西Brazil |
Table 6 The hosts and geographical regions of recombinant parents of ASGV-tree peony isolates
| 序列 Sequence | 分离物 Isolate | 寄主 Host | 地理位置 Geographical region |
|---|---|---|---|
| OQ658829 | Peony BJ2 | 牡丹Tree peony | 中国China |
| OQ658831 | Peony BJ4 | 牡丹Tree peony | 中国China |
| OQ658832 | Peony BJ5 | 牡丹Tree peony | 中国China |
| OQ658833 | Peony BJ6 | 牡丹Tree peony | 中国China |
| OQ658834 | Peony BJ7 | 牡丹Tree peony | 中国China |
| OQ658835 | Peony BJ8 | 牡丹Tree peony | 中国China |
| MT607622 | Peony BJ | 牡丹Tree peony | 中国China |
| MK929792 | BR-Brael | 苹果Apple | 巴西Brazil |
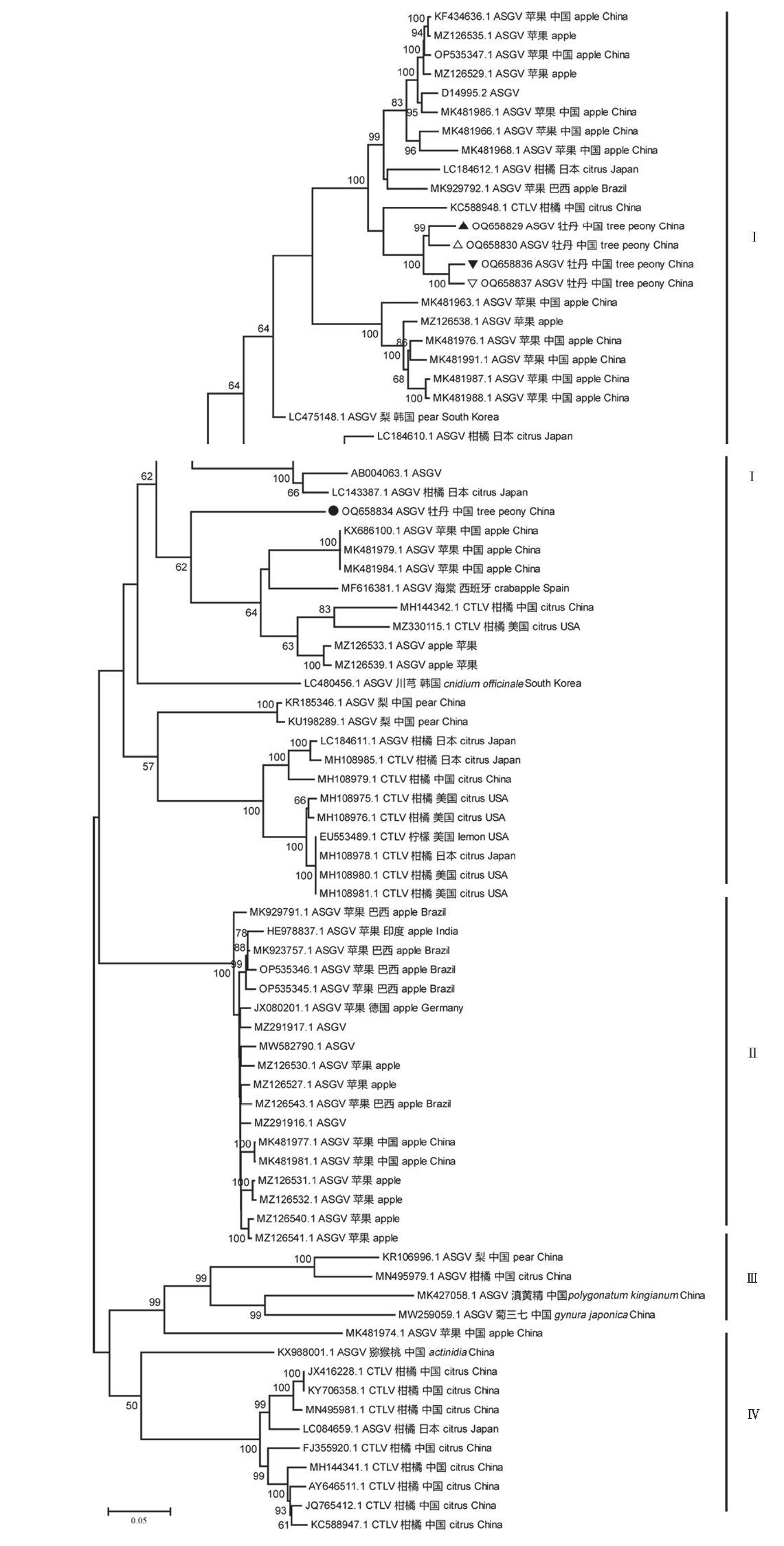
Fig. 4 Phylogenetic analysis of the apple stem grooving virus(ASGV)based on all available non-recombinant complete genome sequences The complete genome sequences of five non-recombinant ASGV-tree peony isolates labeled with triangle and circle.
| [1] |
|
| [2] |
doi: 10.1099/jgv.0.001142 pmid: 30156526 |
| [3] |
doi: 10.1007/s10658-018-1535-3 |
| [4] |
doi: 10.1046/j.1365-3059.2003.00857.x URL |
| [5] |
doi: 10.1007/s10327-022-01051-y |
| [6] |
|
| [7] |
|
| [8] |
doi: 10.1099/vir.0.19179-0 URL |
| [9] |
doi: 10.3186/jjphytopath.45.712 URL |
| [10] |
doi: 10.1007/s10327-021-01039-0 |
| [11] |
|
| [12] |
doi: 10.1094/PDIS-01-21-0007-RE pmid: 34156278 |
| [13] |
|
|
贾晓君, 胡国君, 张尊平, 范旭东, 任芳, 董雅凤. 2023. 苹果茎沟病毒侵染对嘎拉苹果果实品质的影响. 中国果树,(3):18-22.
|
|
| [14] |
|
| [15] |
pmid: 17298878 |
| [16] |
doi: 10.1021/jf902103b URL |
| [17] |
doi: 10.1007/s00239-012-9518-z pmid: 23149596 |
| [18] |
doi: 10.1007/s41348-022-00665-w |
| [19] |
doi: 10.1099/vir.0.82335-0 pmid: 17170463 |
| [20] |
doi: 10.1111/jph.v169.11-12 URL |
| [21] |
|
| [22] |
|
| [23] |
doi: 10.1093/molbev/mst197 URL |
| [24] |
|
| [25] |
doi: 10.1094/PHYTO-99-4-0423 URL |
| [26] |
doi: 10.1080/07060661.2017.1401006 URL |
| [27] |
|
|
谢利媛, 王瑞群. 2022. 菏泽牡丹产业十年蝶变. 中国花卉报,1-5.
|
|
| [28] |
doi: 10.1016/S2095-3119(21)63823-6 URL |
| [29] |
doi: 10.1099/0022-1317-74-12-2743 URL |
| [30] |
pmid: 1413530 |
| [1] | KUANG Xuekun, ZHENG Qian, KONG Qingbo, ZHOU Jiasi, ZHOU Lijun, FENG Shiling, DING Chunbang, and CHEN Tao . Research Progress on Phylogeny and Classification of Camellia [J]. Acta Horticulturae Sinica, 2024, 51(1): 53-66. |
| [2] | KANG Xiaofei , LIU Yumei , ZHAO Chunlei , WANG Bo , and LI Xia, . A New Tree Peony Cultivar‘Lan Xi’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 147-148. |
| [3] | NIE Xinghua, ZHANG Yu, LIU Song, YANG Jiabin, HAO Yaqiong, LIU Yang, QIN Ling, XING Yu. Study on Genetic Characteristics and Taxonomic Status of Wild Chinese Chestnut Based on Genome Re-sequencing [J]. Acta Horticulturae Sinica, 2023, 50(8): 1622-1636. |
| [4] | XUE Yuqian, LIU Zhiyong, SUN Kairong, ZHANG Xiuxin, LÜ Yingmin, XUE Jingqi. The Mechanism of Sugar Signal Involved in Regulating Re-flowering of Tree Peony Under Forcing Culture [J]. Acta Horticulturae Sinica, 2023, 50(3): 596-606. |
| [5] | LI Ruiya, SONG Chengwei, NIU Tongfei, WEI Zhenzhen, GUO Lili, HOU Xiaogai. The Emitted Pattern Analysis of Flower Volatiles and Cloning of PsGDS Gene in Tree Peony Cultivar‘High Noon’ [J]. Acta Horticulturae Sinica, 2023, 50(2): 331-344. |
| [6] | HE Chengyong, ZHAO Xiaoli, XU Tengfei, GAO Dehang, LI Shifang, WANG Hongqing. Genome Sequence Analysis of Strawberry Virus 1 from Shandong Province,China [J]. Acta Horticulturae Sinica, 2023, 50(1): 153-160. |
| [7] | HE Zhihong, HE Lixia, ZHANG Yandong, YANG Guozhou, LI Rui, XU Jingjing, QU Dan, and LI Jingjing. A New Tree Peony Cultivar‘Yuxia Sanqi’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 219-220. |
| [8] | ZHAO Haijun, GAI Shupeng, CHAO Zhen, YAN Shanshan, FANG Yifu, and ZHANG Pei. A New Tree Peony Cultivar‘Fu Zhao Fen Lan’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 221-222. |
| [9] | DING Zhijie, BAO Jinbo, ROUXIAN Guli, ZHU Tiantian, LI Xueli, MIAO Haoyu, TIAN Xinmin. Comparative Chloroplast Genome Study of Mallus servisii‘Red Delicious’and‘Golden Delicious’ [J]. Acta Horticulturae Sinica, 2022, 49(9): 1977-1990. |
| [10] | JIANG Sisi, YUAN Jun, ZHOU Wenjun, NIU Genhua, ZHOU Junqin. Complete Chloroplast Genome Sequence and Characteristics Analysis of Carya illinoinensis [J]. Acta Horticulturae Sinica, 2022, 49(8): 1772-1784. |
| [11] | ZHONG Jing, ZHAO Liling, LI Tingting, ZHANG Shuiying, CHEN Yue, DING Ming. Molecular Identification and Sequence Analysis of Begomoviruses Infecting Impatiens balsamina [J]. Acta Horticulturae Sinica, 2022, 49(5): 1136-1144. |
| [12] | LI Shifan, SHAO Yuchun, QI Haonan, YANG Lan, LI Xiaowei, XU Bingliang, CHEN Yahan. Detection and Genetic Diversity Analysis of Apple Necrotic Mosaic Virus in Gansu Province [J]. Acta Horticulturae Sinica, 2022, 49(11): 2431-2438. |
| [13] | LIU Rong, CI Huiting, REN Xiuxia, GAO Jie, WANG Shunli, ZHANG Xiuxin. Optimization of Callus Induction from Immature Embryo and Establishment Regeneration System of Paeonia ostii‘Fengdan’ [J]. Acta Horticulturae Sinica, 2022, 49(1): 166-174. |
| [14] | YE Kang, HU Yonghong, and ZHANG Ying. A New Tree Peony Cultivar‘Jinliu Hewu’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2959-2960. |
| [15] | YE Kang, HU Yonghong, and ZHANG Ying. A New Tree Peony Cultivar‘Yinsu Ziran’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2961-2962. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd