
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (7): 1329-1339.doi: 10.16420/j.issn.0513-353x.2020-1032
• Research Papars • Previous Articles Next Articles
CHEN Bin, WU Zhen, WEN Junqin, LIN Haowei, YU Lu, XUE Lingzi, ZHOU Rong, JIANG Fangling*( )
)
Received:2021-02-07
Revised:2021-05-25
Online:2021-07-25
Published:2021-08-10
Contact:
JIANG Fangling
E-mail:jfl@njau.edu.cn
CLC Number:
CHEN Bin, WU Zhen, WEN Junqin, LIN Haowei, YU Lu, XUE Lingzi, ZHOU Rong, JIANG Fangling. QTL Mapping and Candidate Genes Analysis of Irregular Fruit Cracking in Tomato[J]. Acta Horticulturae Sinica, 2021, 48(7): 1329-1339.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-1032
| 标记类型 | 标记 | 正向引物(5′-3′) | 反向引物(5′-3′) |
|---|---|---|---|
| Protocol | Marker | Forward primer | Reverse primer |
| SSR | SSR50 | CCGTGACCCTCTTTACAAGC | TTGCTTTCTTCTTCGCCATT |
| SSR287 | GCATCCCAAACAATCCAATC | TCCACTTTCAAGATCAGAGCAA | |
| TES373 | GCCAATGGCTTTGTTGAAGGT | ATGGTGACTTTGCCCTTGTC | |
| TES992 | GATGGACTGGCTCTGGCTCTA | TCACCATCACTGCTTCCATC | |
| InDel | C02M36734 | GACAAATATCATTCGTTTTAATG | CCAAAACACCAAATAATTACTAA |
| C02M52141 | TTCTTATGTCCTCCGAACAAC | GGACTACACCCTACTTAGGCTA | |
| Bin3361 | CAGGGGAACTCCAAATTTCA | CACATGGTATCAAAGCACGG | |
| Bin3371 | GAAGTTGACACAATCCCGCT | TCGAAATCAGATCCAACTGC | |
| sli2734 | GGTAGGGATCTTGAGCATGG | GGAGCCAGAGGGATCAAAAG | |
| sli2763 | AAGGTCTCTCCCTTTGAGCTG | TTGATCATGAGGGTAGCCTTG |
Table 1 Primer sequences of partial markers
| 标记类型 | 标记 | 正向引物(5′-3′) | 反向引物(5′-3′) |
|---|---|---|---|
| Protocol | Marker | Forward primer | Reverse primer |
| SSR | SSR50 | CCGTGACCCTCTTTACAAGC | TTGCTTTCTTCTTCGCCATT |
| SSR287 | GCATCCCAAACAATCCAATC | TCCACTTTCAAGATCAGAGCAA | |
| TES373 | GCCAATGGCTTTGTTGAAGGT | ATGGTGACTTTGCCCTTGTC | |
| TES992 | GATGGACTGGCTCTGGCTCTA | TCACCATCACTGCTTCCATC | |
| InDel | C02M36734 | GACAAATATCATTCGTTTTAATG | CCAAAACACCAAATAATTACTAA |
| C02M52141 | TTCTTATGTCCTCCGAACAAC | GGACTACACCCTACTTAGGCTA | |
| Bin3361 | CAGGGGAACTCCAAATTTCA | CACATGGTATCAAAGCACGG | |
| Bin3371 | GAAGTTGACACAATCCCGCT | TCGAAATCAGATCCAACTGC | |
| sli2734 | GGTAGGGATCTTGAGCATGG | GGAGCCAGAGGGATCAAAAG | |
| sli2763 | AAGGTCTCTCCCTTTGAGCTG | TTGATCATGAGGGTAGCCTTG |
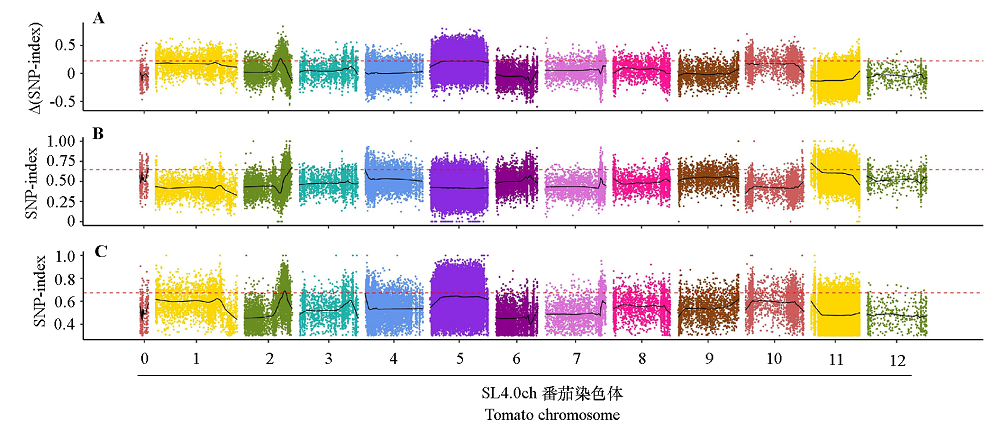
Fig. 3 Distribution of Δ(SNP-index)(A),WB-SNP-index(B)and MB-SNP-index(C)in the whole genome The scattered points in the figure represent the calculated values of ΔSNP-index and SNP-index;The black curve represents the corresponding fitting value;The red dotted line represents 99% threshold line set by LOESS.
| [1] |
Abbott J D, Peet M M, Willits D H, Sanders D C, Gough R E. 1986. Effects of irrigation frequency and scheduling on fruit production and radial fruit cracking in greenhouse tomatoes in soil beds and in a soil-less medium in bags. Scientia Horticulturae, 28(3):209-217.
doi: 10.1016/0304-4238(86)90002-6 URL |
| [2] |
Capel C, Yuste-Lisbona F J, Lopez-Casado G, Angosto T, Cuartero J, Lozano R, Capel J. 2017. Multi-environment QTL mapping reveals genetic architecture of fruit cracking in a tomato RIL Solanum lycopersicum × S-pimpinellifolium population. Theoretical and Applied Genetics, 130(1):213-222.
doi: 10.1007/s00122-016-2809-9 URL |
| [3] |
Cuartero J, Palomares G, Balasch S, Nuez, F. 1981. Tomato fruit cracking under plastic-house and in the open air. ii. general and specific combining abilities. BMC Infectious Diseases, 15(1):1-13.
doi: 10.1186/s12879-014-0722-x URL |
| [4] |
Gine-Bordonaba J, Echeverria G, Ubach D, Aguilo-Aguayo I, Lopez M L, Larrigaudiere C. 2017. Biochemical and physiological changes during fruit development and ripening of two sweet cherry varieties with different levels of cracking tolerance. Plant Physiology and Biochemistry, 111:216-225.
doi: 10.1016/j.plaphy.2016.12.002 URL |
| [5] |
Hovav R, Chehanovsky N, Moy M, Jetter R, Schaffer A A. 2007. The identification of a gene(Cwp1),silenced during Solanum evolution,which causes cuticle microfissuring and dehydration when expressed in tomato fruit. Plant Journal, 52(4):627-639.
pmid: 17877702 |
| [6] | Hudson I W. 1956. The inheritance of resistance to fruit cracking in the tomato(Lycopersicon esculentum L.)[M. D. Dissertation]. Corvallis,US:Oregon State University. |
| [7] |
Jiang F L, Lopez A, Jeon S, de Freitas S T, Yu Q H, Wu Z, Labavitch J M, Tian S K, Powell A L T, Mitcham E. 2019. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Horticulture Research, 6:17.
doi: 10.1038/s41438-018-0105-3 URL |
| [8] |
Kadambari G, Vemireddy L R, Srividhya A, Nagireddy R, Jena S S, Gandikota M, Patil S, Veeraghattapu R, Deborah D A K, Reddy G E, Shake M, Dasari A, Ramanarao P V, Durgarani C V, Neeraja C N, Siddiq E A, Sheshumadhav M. 2018. QTL-Seq-based genetic analysis identifies a major genomic region governing dwarfness in rice(Oryza sativa L.). Plant Cell Reports, 37(4):677-687.
doi: 10.1007/s00299-018-2260-2 URL |
| [9] |
Khadivi-Khub A. 2015. Physiological and genetic factors influencing fruit cracking. Acta Physiologiae Plantarum, 37(1):1718.
doi: 10.1007/s11738-014-1718-2 URL |
| [10] |
Kunwar S, Iriarte F, Fan Q R, da Silva E E, Ritchie L, Nguyen N S, Freeman J H, Stall R E, Jones J B, Minsavage G V, Colee J, Scott J W, Vallad G E, Zipfel C, Horvath D, Westwood J, Hutton S F, Paret M L. 2018. Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology, 108(12):1402-1411.
doi: 10.1094/PHYTO-12-17-0424-R pmid: 29923802 |
| [11] |
Leclercq J, Adams-Phillips L C, Zegzouti H, Jones B, Bouzayen M. 2002. Lectr1,a tomato ctr1-like gene,demonstrates ethylene signaling ability in Arabidopsis and novel expression patterns in tomato. Plant Physiology, 130(3):1132-1142.
pmid: 12427980 |
| [12] | Li Hui-jia. 2016. Genetic analysis and QTL mapping in tomato cracking[M. D. Dissertation]. Harbin:Northeast Agricultural University. (in Chinese) |
| 李会佳. 2016. 番茄裂果性状的遗传分析及QTL定位[硕士论文]. 哈尔滨:东北农业大学. | |
| [13] |
Li R, Sun S, Wang H J, Wang K T, Yu H, Zhou Z, Xin P Y, Chu J F, Zhao T M, Wang H Z, Li J Y, Cui X. 2020. FIS1 encodes a GA2-oxidase that regulates fruit firmness in tomato. Nature Communications, 11(1):5844.
doi: 10.1038/s41467-020-19705-w URL |
| [14] |
Liao N Q, Hu Z Y, Li Y Y, Hao J F, Chen S N, Xue Q, Ma Y Y, Zhang K J, Mahmoud A, Ali A, Malangisha G K, Lyu X L, Yang J H, Zhang M F. 2020. Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biotechnology Journal, 18(4):1066-1077.
doi: 10.1111/pbi.v18.4 URL |
| [15] | Liu Qiang, Zhang Gui-you, Kazuo S. 2000. The plant mitogen-activated protein(MAP)kinase. Acta Botanica Sinica, 42(7):661-667. (in Chinese) |
| 刘强, 张贵友, Kazuo S. 2000. 植物促分裂原活化蛋白(MAP)激酶. 植物学报, 42(7):661-667. | |
| [16] | Liu Zhong-qi, Xue Jun, Jin Feng-mei, Bai Yan-ling. 2007. Relationoship between fruit crack and fruit skin architecture and their heterosis in tomato(Lycopersicon esculentum Mill.). Acta Agriculturae Boreai-Sinica, 22(3):141-147. (in Chinese) |
| 刘仲齐, 薛俊, 金凤媚, 白艳玲. 2007. 番茄裂果与果皮结构的关系及其杂种优势表现. 华北农学报, 22(3):141-147. | |
| [17] |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25(4):402-408.
pmid: 11846609 |
| [18] |
Lopez-Casado G, Matas A J, Dominguez E, Cuartero J, Heredia A. 2007. Biomechanics of isolated tomato(Solanum lycopersicum L.)fruit cuticles:the role of the cutin matrix and polysaccharides. Journal of Experimental Botany, 58(14):3875-3883.
doi: 10.1093/jxb/erm233 URL |
| [19] |
Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, Sun J, Zhang Z, Weng Y, Huang S. 2014. QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theoretical and Applied Genetics, 127(7):1491-1499.
doi: 10.1007/s00122-014-2313-z URL |
| [20] |
Lu K, Li T, He J, Chang W, Zhang R, Liu M, Yu M N, Fan Y H, Ma J Q, Sun W, Qu C M, Liu L Z, Li N N, Liang Y, Wang R, Qian W, Tang Z L, Xu X F, Lei B, Zhang K, Li J A. 2018. qPrimerDB:a thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Research, 46(D1):D1229-D1236.
doi: 10.1093/nar/gkx725 URL |
| [21] |
Moctezuma E, Smith D L, Gross K C. 2003. Antisense suppression of a beta-galactosidase gene(TBG6)in tomato increases fruit cracking. Journal of Experimental Botany, 54(390):2025-2033.
pmid: 12867545 |
| [22] |
Murray M, Thompson W F. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8(19):4321-4326.
pmid: 7433111 |
| [23] |
Pandey M K, Khan A W, Singh V K, Vishwakarma M K, Shasidhar Y, Kumar V, Garg V, Bhat R S, Chitikineni A, Janila P, Guo B, Varshney R K. 2017. QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut(Arachis hypogaea L.). Plant Biotechnology Journal, 15(8):927-941.
doi: 10.1111/pbi.2017.15.issue-8 URL |
| [24] |
Peet M M. 1992. Fruit cracking in tomato. Horttechnology, 2(2):216-223.
doi: 10.21273/HORTTECH.2.2.216 URL |
| [25] |
Singh V K, Khan A W, Jaganathan D, Thudi M, Roorkiwal M, Takagi H, Garg V, Kumar V, Chitikineni A, Gaur P M, Sutton T, Terauchi R, Varshney R K. 2016. QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnology Journal, 14(11):2110-2119.
doi: 10.1111/pbi.2016.14.issue-11 URL |
| [26] |
Sonderby I E, Geu-Flores F, Halkier B A. 2010. Biosynthesis of glucosinolates-gene discovery and beyond. Trends in Plant Science, 15(5):283-290.
doi: 10.1016/j.tplants.2010.02.005 URL |
| [27] | Song Dong-hui, Song Feng-ming, Zheng Zhong. 2004. Function of MAP kinases in plant signal transduction pathway networks. Journal of Zhejiang University(Agriculture and Life Sciences), 30(2):119-126. (in Chinese) |
| 宋东辉, 宋凤鸣, 郑重. 2004. MAP激酶在植物信号传递网络中的功能. 浙江大学学报(农业与生命科学版), 30(2):119-126. | |
| [28] | Tong V G. 2016. Development of InDel markers and fine-mapping of locule number 2.2 in tomato[Ph. D. Dissertation]. Beijing:Chinese Academy of Agricultural Sciences. (in Chinese) |
| Tong Van Giang. 2016. 番茄InDel分子标记的开发与果实心室数量主效QTL lcn2.2的精细定位[博士论文]. 北京:中国农业科学院. | |
| [29] |
Walter J M. 1967. Hereditary resistance to disease in tomato. Annual Review of Phytopathology, 5(1):131-160.
doi: 10.1146/annurev.py.05.090167.001023 URL |
| [30] |
Wang J G, Gao X M, Ma Z L, Chen J, Liu Y N, Shi W Q. 2019. Metabolomic and transcriptomic profiling of three types of litchi pericarps reveals that changes in the hormone balance constitute the molecular basis of the fruit cracking susceptibility of Litchi chinensis cv. Baitangying. Molecular Biology Reports, 46(5):5295-5308.
doi: 10.1007/s11033-019-04986-2 URL |
| [31] | Wei Kai, Liu Xiaoyan, Cao Xue, Liu Xiaolin, Wang Xiaotian, Yang Mengxia, Wang Jing, Wang Xiaoxuan, Guo Yanmei, Du Yongchen, Li Junming, Liu Lei, Shu Jinshuai, Qin Yong, Huang Zejun. 2020. Identification of quantitative trait loci controlling tomato fruit weight by QTL-seq. Acta Horticulturae Sinica, 47(3):571-580. (in Chinese) |
| 魏凯, 刘晓燕, 曹雪, 刘晓林, 王晓甜, 杨孟霞, 王净, 王孝宣, 国艳梅, 杜永臣, 李君明, 刘磊, 舒金帅, 秦勇, 黄泽军. 2020. 利用QTL-seq定位番茄果实质量QTL. 园艺学报, 47(3):571-580. | |
| [32] |
Wen J Q, Jiang F L, Weng Y Q, Sun M T, Shi X P, Zhou Y Z, Yu L, Wu Z. 2019. Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping,QTL-seq and RNA-seq in tomato. BMC Plant Biology, 19(1):398.
doi: 10.1186/s12870-019-2008-3 URL |
| [33] |
Xue L Z, Sun M T, Wu Z, Yu L, Yu Q H, Tang Y P, Jiang F L. 2020. LncRNA regulates tomato fruit cracking by coordinating gene expression via a hormone-redox-cell wall network. BMC Plant Biology, 20(1):162.
doi: 10.1186/s12870-020-02373-9 URL |
| [34] |
Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology, 57:781-803.
pmid: 16669782 |
| [35] |
Yang J J, Wang Y Y, Shen H L, Yang W C. 2014. In silico identification and experimental validation of insertion-deletion polymorphisms in tomato genome. DNA Research, 21(4):429-438.
doi: 10.1093/dnares/dsu008 URL |
| [36] | Yang Ze-en. 2015. Screening of agronomic characters related to tomato fruit cracking and the study on mechanism of tomato fruit cracking based on the pericarp[M. D. Dissertation]. Nanjing:Nanjing Agricultural University. (in Chinese) |
| 杨泽恩. 2015. 番茄裂果相关指标筛选及基于果皮特征的裂果机理研究[硕士论文]. 南京:南京农业大学. | |
| [37] | Zhu Yu, Fan Lina, Huang Zejun, Du Yongchen, Guo Yanmei, Li Junming, Liu Lei, Shu Jinshuai, Wang Xiaoxuan. 2020. Fine mapping of fruit cracking-resistant gene Cr3a in tomato. Acta Horticulturae Sinica, 47(2):275-286. (in Chinese) |
| 朱玉, 范丽娜, 黄泽军, 杜永臣, 国艳梅, 李君明, 刘磊, 舒金帅, 王孝宣. 2020. 番茄耐裂果基因Cr3a的精细定位. 园艺学报, 47(2):275-286. |
| [1] | SHI Hongli, LI La, GUO Cuimei, YU Tingting, JIAN Wei, YANG Xingyong. Isolation,Identification and Analysis of Biocontrol Ability of Biocontrol Strain TL1 Against Tomato Botrytis cinerea [J]. Acta Horticulturae Sinica, 2023, 50(1): 79-90. |
| [2] | HU Jingyu, QUE Kaijuan, MIAO Tianli, WU Shaozheng, WANG Tiantian, ZHANG Lei, DONG Xian, JI Pengzhang, DONG Jiahong. Identification of Tomato Spotted Wilt Orthotospovirus Infecting Iris tectorum [J]. Acta Horticulturae Sinica, 2023, 50(1): 170-176. |
| [3] | ZHENG Jirong, WANG Tonglin, and HU Songshen. A New Tomato Cultivar‘Hangza 603’with High Quality [J]. Acta Horticulturae Sinica, 2022, 49(S2): 103-104. |
| [4] | ZHENG Jirong and WANG Tonglin. A New Tomato Cultivar‘Hangza 601’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 105-106. |
| [5] | ZHENG Jirong and WANG Tonglin. A New Cherry Tomato Cultivar‘Hangza 503’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 107-108. |
| [6] | HUANG Tingting, LIU Shuqin, ZHANG Yongzhi, LI Ping, ZHANG Zhihuan, and SONG Libo. A New Cherry Tomato Cultivar‘Yingshahong 4’ [J]. Acta Horticulturae Sinica, 2022, 49(S2): 109-110. |
| [7] | ZHANG Qianrong, LI Dazhong, QIU Boyin, LIN Hui, MA Huifei, YE Xinru, LIU Jianting, ZHU Haisheng, and WEN Qingfang. A New Tomato Cultivar‘Minnongke 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 73-74. |
| [8] | HAN Shuai, WU Jie, ZHANG Heqing, XI Yadong. Identification and Sequence Analysis of Tomato Spotted Wilt Orthotospovirus Infecting Lettuce in Sichuan [J]. Acta Horticulturae Sinica, 2022, 49(9): 2007-2016. |
| [9] | CHEN Lilang, YANG Tianzhang, CAI Ruping, LIN Xiaoman, DENG Nankang, CHE Haiyan, LIN Yating, KONG Xiangyi. Molecular Detection and Identification of Viruses from Passiflora edulis in Hainan [J]. Acta Horticulturae Sinica, 2022, 49(8): 1785-1794. |
| [10] | LU Tao, YU Hongjun, LI Qiang, JIANG Weijie. Effects of Leaf and Fruit Quantity Regulation on Growth,Fruit Quality and Yield of Tomato [J]. Acta Horticulturae Sinica, 2022, 49(6): 1261-1274. |
| [11] | MENG Xianmin, CUI Qingqing, DUAN Yundan, ZHUANG Tuanjie, PU Dan, DONG Chunjuan, YANG Wencai, SHANG Qingmao. Promoting Effects of Uniconazole on Grafting Formation of Tomato Seedlings and Underlying Mechanisms [J]. Acta Horticulturae Sinica, 2022, 49(6): 1275-1289. |
| [12] | CUI Dongyu, LI Changqing, SUN Yanxin, WANG Jiqing, ZOU Guoyuan, YANG Jungang. Effects of Dwarf Close Planting on Growth and Yield of Tomato Under East-West Cultivation in Greenhouse [J]. Acta Horticulturae Sinica, 2022, 49(4): 875-884. |
| [13] | CHEN Tongqiang, ZHANG Tianzhu, WANG Xiaozhuo. Research Progress of The Regulation of Light on Lycopene Biosynthesis in Tomato Fruit [J]. Acta Horticulturae Sinica, 2022, 49(4): 907-923. |
| [14] | PENG Yi, LI Yuanhui, YANG Rui, ZHANG Ziyi, LI Yanan, HAN Yunhao, ZHAO Wenchao, WANG Shaohui. The Jasmonic Acid Synthesis Gene LoxD Participates in the Regulation of Tomato Drought Resistance [J]. Acta Horticulturae Sinica, 2022, 49(2): 319-331. |
| [15] | FANG Tian, LIU Jihong. Advances in Identification of QTLs Associated with Significant Traits in Major Fruit Trees [J]. Acta Horticulturae Sinica, 2022, 49(12): 2622-2640. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd