
园艺学报 ›› 2025, Vol. 52 ›› Issue (4): 1051-1064.doi: 10.16420/j.issn.0513-353x.2024-0361
收稿日期:2024-11-26
修回日期:2025-01-17
出版日期:2025-05-08
发布日期:2025-04-25
通讯作者:
基金资助:
YUAN Luming, ZHU Hongju, LIU Wenge*( )
)
Received:2024-11-26
Revised:2025-01-17
Published:2025-05-08
Online:2025-04-25
Contact:
摘要:
果肉颜色是园艺作物果实的重要性状,对自身种子传播、营养物质评价,消费者选择都十分重要。果肉颜色表现出多样性及复杂性,比如猕猴桃果肉存在绿、黄、红心,西瓜果肉存在黄、红、白、混合等多种颜色。果肉颜色的形成主要受色素类物质的组成及含量影响,如类胡萝卜素、叶绿素、花青素苷等。通过调控色素类物质代谢通路中的关键基因表达,可以实现对园艺产品颜色的调控。转录因子(Transcription factor,TF)通过与相关基因的启动子区域结合,激活或抑制基因的表达,实现对基因表达和颜色的调控。在前人的研究中,多种转录因子被发掘并证明对色素代谢通路中相关基因及颜色具有重要调控作用,如MYB、NAC、MADS、WRKY等。
袁路明, 朱红菊, 刘文革. 转录因子对园艺作物果肉颜色的调控[J]. 园艺学报, 2025, 52(4): 1051-1064.
YUAN Luming, ZHU Hongju, LIU Wenge. The Regulation of Flesh Color of Horticultural Crops by Transcription Factors[J]. Acta Horticulturae Sinica, 2025, 52(4): 1051-1064.
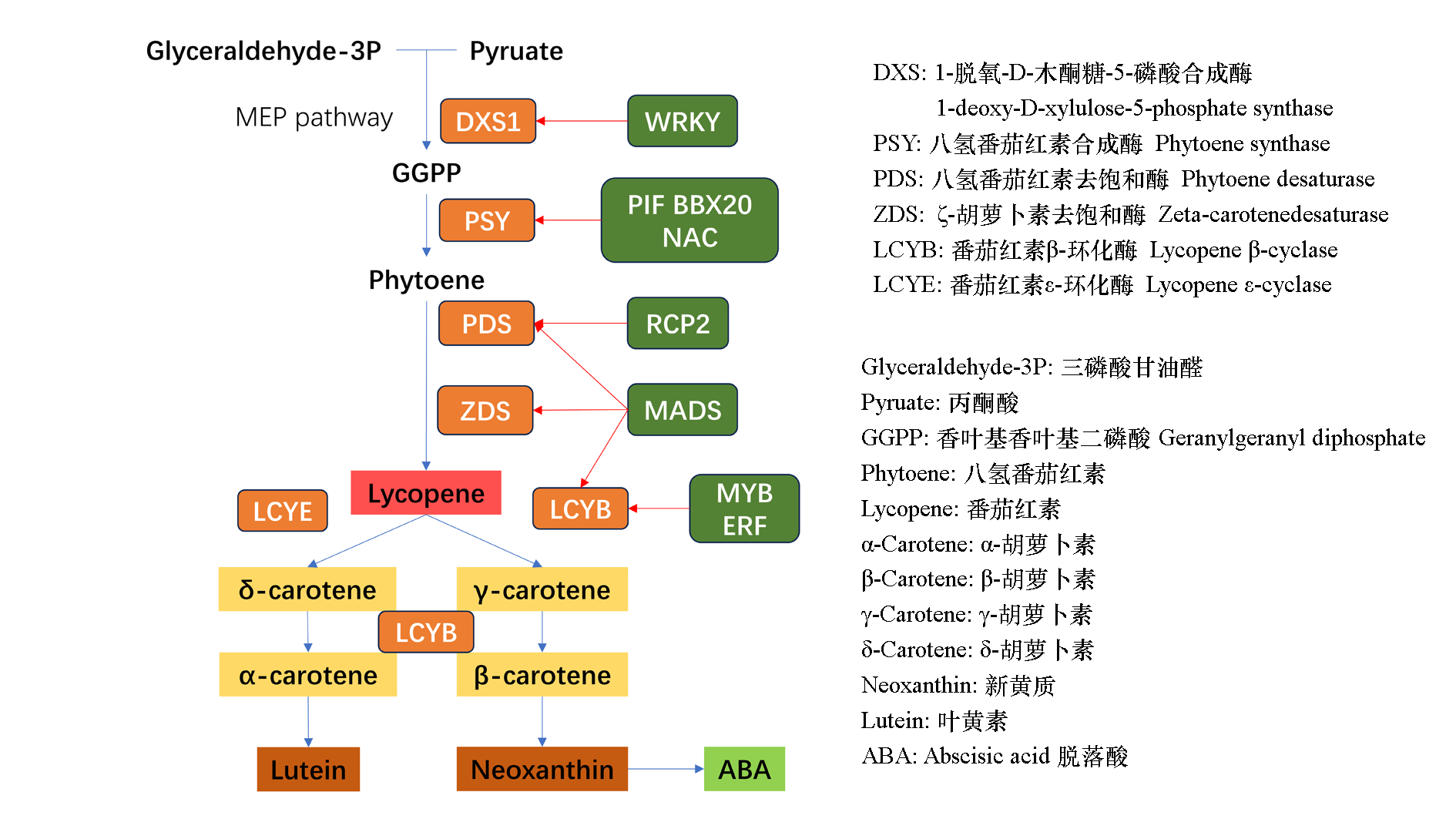
图1 类胡萝卜素代谢通路 蓝色线条代表代谢方向,红色线条代表转录因子参与对相关酶基因的调控,图片形式引自Yuan等(2023),下同
Fig. 1 Carotenoid metabolism pathway Bule lines indicate metabolic direction,red lines indicate transcription factors involved in regulation of related enzyme genes,the format of pictures were cited from Yuan et al.(2023),the following pictures is the same
| 功能 Function | 步骤 Step | 酶 Enzyme | 基因 Gene |
|---|---|---|---|
| 合成 Synthesis | 1 | 谷氨酰-tRNA合成酶 Glutamyl-tRNA synthase | GluRS |
| 2 | 谷氨酰-tRNA还原酶 Glutamyl-tRNA reductase | GluTR | |
| 3 | 谷氨酸-1-半醛转氨酶 Glutamate-1-semialdehyde aminotransferase | GSA-AT | |
| 4 | δ-氨基酮戊酸脱氢酶 δ-Aminolevulinic acid dehydratase | ALAD | |
| 5 | 胆色原素脱氨酶 Porphobilinogen deaminase | PBD | |
| 6 | 尿卟啉原Ⅲ合酶 Uroporphyrinogen III synthase | UroS | |
| 7 | 尿卟啉原Ⅲ脱氢酶 Uroporphyrinogen III dehydrogenase | UroD | |
| 8 | 粪卟啉原氧化酶 Coproporphyrinogen III oxidase | CPO | |
| 9 | 原卟啉原氧化酶 Protoporphyrinogen IX oxidase | PPX | |
| 10 | 铁螯合酶 Ferrochelatase | FeCh | |
| 11 | 镁螯合酶 Magnesium chelatase | MgCh | |
| 12 | Mg-原卟啉Ⅸ甲基转移酶 Mg-protoporphyrin IX methyltransferase | MTF | |
| 13 | Mg-原卟啉Ⅸ单甲酯环化酶 Mg-protoporphyrin IX monomethyl ester cyclase | MTC | |
| 14 | 8-乙烯基还原酶 8-vinyl reductase | VR | |
| 15 | NADPH-原叶绿素酸酯氧化还原酶 NADPH-protochlorophyllide oxidoreductase | POR | |
| 16 | 叶绿素酸酯a加氧酶 Chlorophyllide a oxygenase | CAO | |
| 17 | 叶绿素合酶 Chlorophyll synthase | CHLS | |
| 18 | 叶绿素降解酶 Chlorophyllase | CLH | |
| 降解Degradation | 1 | 永绿基因 NON-YELLOW COLORING1 | NYC1 |
| 2 | 永绿基因类似基因 NYC1-LIKE | NOL | |
| 3 | 羟基叶绿素a还原酶 Hydroxychlorophyll a reductase | HCAR | |
| 4 | 脱镁螯合酶 STAY GREEN | SGR | |
| 5 | 脱镁叶绿素酶 Pheophytinase | Pao | |
| 6 | 脱镁叶绿酸a氧化酶 Pheophorbide a oxygenase | PAO | |
| 7 | RCC还原酶 RCC reductase | RCCR |
表1 高等植物叶绿素合成和降解通路相关酶及其对应的基因
Table 1 enzymes and corresponding genes in the chlorophyll synthesis and degradation pathway of higher plant
| 功能 Function | 步骤 Step | 酶 Enzyme | 基因 Gene |
|---|---|---|---|
| 合成 Synthesis | 1 | 谷氨酰-tRNA合成酶 Glutamyl-tRNA synthase | GluRS |
| 2 | 谷氨酰-tRNA还原酶 Glutamyl-tRNA reductase | GluTR | |
| 3 | 谷氨酸-1-半醛转氨酶 Glutamate-1-semialdehyde aminotransferase | GSA-AT | |
| 4 | δ-氨基酮戊酸脱氢酶 δ-Aminolevulinic acid dehydratase | ALAD | |
| 5 | 胆色原素脱氨酶 Porphobilinogen deaminase | PBD | |
| 6 | 尿卟啉原Ⅲ合酶 Uroporphyrinogen III synthase | UroS | |
| 7 | 尿卟啉原Ⅲ脱氢酶 Uroporphyrinogen III dehydrogenase | UroD | |
| 8 | 粪卟啉原氧化酶 Coproporphyrinogen III oxidase | CPO | |
| 9 | 原卟啉原氧化酶 Protoporphyrinogen IX oxidase | PPX | |
| 10 | 铁螯合酶 Ferrochelatase | FeCh | |
| 11 | 镁螯合酶 Magnesium chelatase | MgCh | |
| 12 | Mg-原卟啉Ⅸ甲基转移酶 Mg-protoporphyrin IX methyltransferase | MTF | |
| 13 | Mg-原卟啉Ⅸ单甲酯环化酶 Mg-protoporphyrin IX monomethyl ester cyclase | MTC | |
| 14 | 8-乙烯基还原酶 8-vinyl reductase | VR | |
| 15 | NADPH-原叶绿素酸酯氧化还原酶 NADPH-protochlorophyllide oxidoreductase | POR | |
| 16 | 叶绿素酸酯a加氧酶 Chlorophyllide a oxygenase | CAO | |
| 17 | 叶绿素合酶 Chlorophyll synthase | CHLS | |
| 18 | 叶绿素降解酶 Chlorophyllase | CLH | |
| 降解Degradation | 1 | 永绿基因 NON-YELLOW COLORING1 | NYC1 |
| 2 | 永绿基因类似基因 NYC1-LIKE | NOL | |
| 3 | 羟基叶绿素a还原酶 Hydroxychlorophyll a reductase | HCAR | |
| 4 | 脱镁螯合酶 STAY GREEN | SGR | |
| 5 | 脱镁叶绿素酶 Pheophytinase | Pao | |
| 6 | 脱镁叶绿酸a氧化酶 Pheophorbide a oxygenase | PAO | |
| 7 | RCC还原酶 RCC reductase | RCCR |
| 主要作物 Crop | 转录因子 TF | 主要靶基因 Target gene | 果肉颜色表现 Flesh color |
|---|---|---|---|
| 番茄 Solanum lycopersicum | SlAN2 | Ethylene-related genes | 橙黄Orange |
| SlMYB12 | Flavonoid-related genes | 多种颜色Muti-colors | |
| SlNAC1 | SlPSY1 & Ethylene-related genes | 黄色或橙黄色Yellow or orange | |
| SlRIN& SLcMYB1 | Muti-genes | 多种颜色Muti-colors | |
| SlWRKY35 | SlDXS1 | 黄色Yellow | |
| SlERF | SlLCYB2 | 黄色或橙黄色Yellow or orange | |
| SlGLK | SlPOR、Photosynthetic-related gene | ||
| 桃 Prunus persica | PpMYB10.1 | PpUFGT,PpDFR | 淡色Pale |
| PbMYB18 | 红色Red | ||
| PbNAC1 & BL | PpMYB10.1 | 红色Red | |
| 猕猴桃 Actinidia chinensis | AdMYB10 & AdMYB110 | Anthocyanin-related genes | 红色Red |
| AdMYB7 | AdLCYB | 橙黄Orange | |
| 柑橘 Citrus | CsMADS3 | CsPSY1,CsLCYB2,CsSGR | 黄色或橙黄色Yellow or orange |
| CsMADS6 | CsPDS,CsCCD1,CsPSY | 黄色或橙黄色Yellow or orange | |
| 梨 Pyrus | PpWRKY44 & PpMYB10 | PpCHS,PpF3H | 红色Red |
| 苹果 Malus | MdMYBDL1 & MdHY5 | MdMYB16 | 红色Red |
| 甜瓜Melo | CmNAC-NOR | CmWINV2,CmACS5,CmNCED3,CmZDS1 | 橙黄Orange |
| CmWRKY49 | CmPSY1 | 橙黄Orange |
表2 主要园艺作物中调控果肉颜色性状的转录因子及其靶基因
Table 2 Transcription factors and target genes during the regulation of flesh color in major horticultural crops
| 主要作物 Crop | 转录因子 TF | 主要靶基因 Target gene | 果肉颜色表现 Flesh color |
|---|---|---|---|
| 番茄 Solanum lycopersicum | SlAN2 | Ethylene-related genes | 橙黄Orange |
| SlMYB12 | Flavonoid-related genes | 多种颜色Muti-colors | |
| SlNAC1 | SlPSY1 & Ethylene-related genes | 黄色或橙黄色Yellow or orange | |
| SlRIN& SLcMYB1 | Muti-genes | 多种颜色Muti-colors | |
| SlWRKY35 | SlDXS1 | 黄色Yellow | |
| SlERF | SlLCYB2 | 黄色或橙黄色Yellow or orange | |
| SlGLK | SlPOR、Photosynthetic-related gene | ||
| 桃 Prunus persica | PpMYB10.1 | PpUFGT,PpDFR | 淡色Pale |
| PbMYB18 | 红色Red | ||
| PbNAC1 & BL | PpMYB10.1 | 红色Red | |
| 猕猴桃 Actinidia chinensis | AdMYB10 & AdMYB110 | Anthocyanin-related genes | 红色Red |
| AdMYB7 | AdLCYB | 橙黄Orange | |
| 柑橘 Citrus | CsMADS3 | CsPSY1,CsLCYB2,CsSGR | 黄色或橙黄色Yellow or orange |
| CsMADS6 | CsPDS,CsCCD1,CsPSY | 黄色或橙黄色Yellow or orange | |
| 梨 Pyrus | PpWRKY44 & PpMYB10 | PpCHS,PpF3H | 红色Red |
| 苹果 Malus | MdMYBDL1 & MdHY5 | MdMYB16 | 红色Red |
| 甜瓜Melo | CmNAC-NOR | CmWINV2,CmACS5,CmNCED3,CmZDS1 | 橙黄Orange |
| CmWRKY49 | CmPSY1 | 橙黄Orange |
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
|
贺威智, 雷伟奇, 郭祥鑫, 李蕊莲, 陈冠群. 2023. 百子莲MYB家族鉴定及蓝色形成关键基因功能分析. 园艺学报, 50 (6):1255-1268.
|
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
| [77] |
|
| [78] |
|
| [1] | 梁国平, 曾宝珍, 刘铭, 边志远, 陈佰鸿, 毛娟. 山葡萄VaSR基因家族的鉴定及VaSR1抗寒功能验证与互作蛋白筛选[J]. 园艺学报, 2025, 52(1): 37-50. |
| [2] | 杨艳, 刘军, 周晓慧, 刘松瑜, 庄勇. 茄子SmWRKY4的克隆及耐冷性功能验证[J]. 园艺学报, 2025, 52(1): 101-110. |
| [3] | 刘建豪, 荆彦付, 刘月芯, 徐摇光, 于洋, 葛秀秀, 谢华. 桃NAC基因家族的鉴定及PpNAC050促进果实果糖积累研究[J]. 园艺学报, 2024, 51(9): 1983-1996. |
| [4] | 段敏杰, 李怡斐, 王春萍, 杨小苗, 黄任中, 黄启中, 张世才. 辣椒果实类胡萝卜素调控因子转录组和靶向代谢组分析[J]. 园艺学报, 2024, 51(8): 1773-1791. |
| [5] | 赵泽阳, 周雨晴, 林德书, 任慧波. 花瓣锥形表皮细胞形态建成研究进展[J]. 园艺学报, 2024, 51(7): 1695-1706. |
| [6] | 邓淑芳, 刘倩, 刘玲, 陈鸥, 王文军, 曾凯芳, 邓丽莉. 蜜橘CcHY5的克隆及其对果实转色功能的研究[J]. 园艺学报, 2024, 51(5): 939-955. |
| [7] | 孙会茹, 党峰峰, 任敏, 张嘉宁, 樊蓓, 陈国梁, 程国亭, 王延峰. 番茄SlWRKY46调控低温胁迫响应的功能研究[J]. 园艺学报, 2024, 51(12): 2758-2774. |
| [8] | 姜波, 吕远达, 刘淑梅, 闫化学. 植物激素调控柑橘果实成熟的研究进展[J]. 园艺学报, 2024, 51(12): 2928-2944. |
| [9] | 唐征, 陈思雀, 徐谦, 钟伟杰, 刘庆, 朱世杨. AP2/ERF在青花菜苗期响应黑腐病的功能研究[J]. 园艺学报, 2024, 51(11): 2523-2539. |
| [10] | 吴丹, 柳佳欣, 卓林熙, 李钰, 罗英, 周勇, 杨有新, 余婷. CaWRKY39在辣椒响应疫霉菌侵染中的功能分析[J]. 园艺学报, 2024, 51(10): 2297-2310. |
| [11] | 尤倩, 刘晓, 刘梦梦, 刘丹, 伯晨, 朱艳芳, 段永波, 薛建平, 张爱民, 薛涛. 半夏热激因子HSF家族基因鉴定及生物信息分析[J]. 园艺学报, 2024, 51(10): 2371-2385. |
| [12] | 翟挺楷, 马湘玮, 陈燕, 张雪莹, 李卓蕴, 徐卢振, 傅卓然, 赖钟雄, 林玉玲. 染色质免疫共沉淀测序技术及其在园艺植物中的应用研究进展[J]. 园艺学报, 2024, 51(1): 39-52. |
| [13] | 张巧丽, 陈笛, 宋艳萍, 朱鸿亮, 罗云波, 曲桂芹. 番茄果实叶绿素代谢转录调控网络研究进展[J]. 园艺学报, 2023, 50(9): 2031-2047. |
| [14] | 肖翔, 周储江, 金舒婉, 施丽愉, 杨震峰, 曹士锋, 陈伟. PpMADS2与PpMADS3协同调控黄肉桃果实类胡萝卜素积累机制的研究[J]. 园艺学报, 2023, 50(6): 1173-1186. |
| [15] | 贺威智, 雷伟奇, 郭祥鑫, 李蕊莲, 陈冠群. 百子莲MYB家族鉴定及蓝色形成关键基因功能分析[J]. 园艺学报, 2023, 50(6): 1255-1268. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司