
园艺学报 ›› 2022, Vol. 49 ›› Issue (6): 1301-1312.doi: 10.16420/j.issn.0513-353x.2021-0629
收稿日期:2021-06-29
修回日期:2021-11-27
出版日期:2022-06-25
发布日期:2022-07-05
通讯作者:
谭晨
E-mail:tanchen2020@gnnu.edu.cn
基金资助:
CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen( )
)
Received:2021-06-29
Revised:2021-11-27
Online:2022-06-25
Published:2022-07-05
Contact:
TAN Chen
E-mail:tanchen2020@gnnu.edu.cn
摘要:
PAP1/2转录因子是参与形成MBW复合体从而调控花青素合成的主要MYB转录因子。以拟南芥PAP1/2转录因子核酸和蛋白序列比对及Pfam验证,研究白菜、甘蓝和甘蓝型油菜等芸薹属植物中PAP1/2转录因子调控花青素代谢的确切同源拷贝数、进化关系及表达情况:在白菜、甘蓝及甘蓝型油菜中鉴定到的PAP1/2转录因子同源拷贝数分别为3、4和9个;通过进化分析和PAP1/2转录因子同源拷贝所在区段微共线性分析进一步明确了其不同拷贝间的进化关系,厘清了A7和C6染色体上PAP1/2转录因子不同拷贝的相互关系;结合白菜、甘蓝和甘蓝型油菜不同组织转录组测序数据进行表达情况分析,阐明了PAP1/2转录因子同源拷贝在不同组织中的表达情况。
中图分类号:
陈道宗, 刘镒, 沈文杰, 朱博, 谭晨. 白菜、甘蓝和甘蓝型油菜PAP1/2同源基因的鉴定及分析[J]. 园艺学报, 2022, 49(6): 1301-1312.
CHEN Daozong, LIU Yi, SHEN Wenjie, ZHU Bo, TAN Chen. Identification and Analysis of PAP1/2 Homologous Genes in Brassica rapa,B. oleracea and B. napus[J]. Acta Horticulturae Sinica, 2022, 49(6): 1301-1312.
| Function | 拟南芥 A. thaliana | 白菜 B. rapa | 甘蓝 B. oleracea | 甘蓝型油菜-AA B. napus-AA | 甘蓝型油菜-CC B. napus-CC |
|---|---|---|---|---|---|
| PAP1(MYB75) | AT1G56650 | BraA02g017040.3C | Bo00835s060.1 | BnaA02G0160600ZS | BnaC02G0205500ZS |
| PAP1(MYB75) | AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS |
| PAP1(MYB75) | AT1G56650 | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| PAP1(MYB75) | AT1G56650 | BnaC06G0328700ZS | |||
| PAP2(MYB90) | AT1G66390 | BraA07g032100.3C | Bo6g100940.1 | BnaA07G0287000ZS | BnaC06G0329100ZS |
表1 拟南芥、白菜、甘蓝和甘蓝型油菜PAP1/2同源基因拷贝数及共线性关系
Table 1 Copy number and collinearity of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus
| Function | 拟南芥 A. thaliana | 白菜 B. rapa | 甘蓝 B. oleracea | 甘蓝型油菜-AA B. napus-AA | 甘蓝型油菜-CC B. napus-CC |
|---|---|---|---|---|---|
| PAP1(MYB75) | AT1G56650 | BraA02g017040.3C | Bo00835s060.1 | BnaA02G0160600ZS | BnaC02G0205500ZS |
| PAP1(MYB75) | AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS |
| PAP1(MYB75) | AT1G56650 | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| PAP1(MYB75) | AT1G56650 | BnaC06G0328700ZS | |||
| PAP2(MYB90) | AT1G66390 | BraA07g032100.3C | Bo6g100940.1 | BnaA07G0287000ZS | BnaC06G0329100ZS |
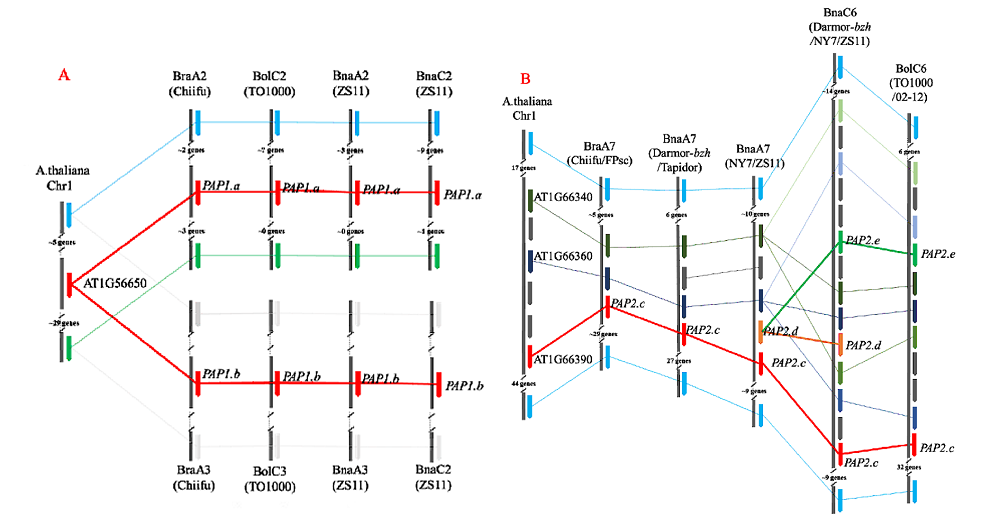
图2 拟南芥、白菜、甘蓝和甘蓝型油菜PAP1/2同源基因所在区段微共线性分析 A. A2-C2、A3-C3染色体PAP1/2同源基因所在区段微共线性分析;B. A7-C6染色体PAP1/2同源基因所在区段微共线性分析。
Fig. 1 The comparison map of the segment of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus A. The comparison map of the segment of PAP1/2 homologous genes on A2-C2,A3-C3 chromosome;B. The comparison map of the segment of PAP1/2 homologous genes on A2-C6 chromosome.
| 拟南芥 A. thaliana | 白菜B. rapa Chiifu-A2 | 甘蓝B. oleracea TO1000-C2 | 甘蓝型油菜B. napus ZS11-A2 | 甘蓝型油菜B. napus ZS11-C2 | |
|---|---|---|---|---|---|
| A2-C2 | AT1G66330 | BraA02g017010.3C | Bo2g052820.1 | BnaA02G0160200ZS | BnaC02T0204500ZS |
| AT1G66390 | BraA02g017040.3C | Bo00835s060.1 | BnaA02T0160600ZS | BnaC02T0205500ZS | |
| AT1G66470 | BraA02g017080.3C | Bo00835s050.1 | BnaA02T0160700ZS | BnaC02T0205700ZS | |
| A3-C3 | AT1G66330 | Null | Null | Null | Null |
| AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS | |
| AT1G66470 | Null | Null | Null | Null | |
| A7-C6 | AT1G66210 | BraA07g032000.3C | Bo6g099780.1 | BnaA07T0285500ZS | BnaC06T0327300ZS |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328700ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0287000ZS | BnaC06G0329100ZS | |
| AT1G66840 | BraA07g032410.3C | Bo6g101320.1 | BnaA07T0289800ZS | BnaC06T0333000ZS |
表2 拟南芥、白菜、甘蓝和甘蓝型油菜PAP1/2同源基因所在区段微共线性分析
Table 2 The comparison map of the segment of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus
| 拟南芥 A. thaliana | 白菜B. rapa Chiifu-A2 | 甘蓝B. oleracea TO1000-C2 | 甘蓝型油菜B. napus ZS11-A2 | 甘蓝型油菜B. napus ZS11-C2 | |
|---|---|---|---|---|---|
| A2-C2 | AT1G66330 | BraA02g017010.3C | Bo2g052820.1 | BnaA02G0160200ZS | BnaC02T0204500ZS |
| AT1G66390 | BraA02g017040.3C | Bo00835s060.1 | BnaA02T0160600ZS | BnaC02T0205500ZS | |
| AT1G66470 | BraA02g017080.3C | Bo00835s050.1 | BnaA02T0160700ZS | BnaC02T0205700ZS | |
| A3-C3 | AT1G66330 | Null | Null | Null | Null |
| AT1G56650 | BraA03g040840.3C | Bo3g081880.1 | BnaA03G0376600ZS | BnaC03G0461400ZS | |
| AT1G66470 | Null | Null | Null | Null | |
| A7-C6 | AT1G66210 | BraA07g032000.3C | Bo6g099780.1 | BnaA07T0285500ZS | BnaC06T0327300ZS |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328400ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0286900ZS | BnaC06G0328700ZS | |
| AT1G66390 | BraA07g032100.3C | Bo6g099880.1 | BnaA07G0287000ZS | BnaC06G0329100ZS | |
| AT1G66840 | BraA07g032410.3C | Bo6g101320.1 | BnaA07T0289800ZS | BnaC06T0333000ZS |

图3 拟南芥、白菜、甘蓝和甘蓝型油菜PAP1/2同源基因R2、R3保守结构域基序
Fig. 3 R2,R3 conserved domain motif of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus

图4 拟南芥、白菜、甘蓝和甘蓝型油菜不同组织PAP1/2同源基因蛋白保守基序
Fig. 4 Protein conservation motif analysis of PAP1/2 homologous genes in different tissues of Arabidopsis,Brassica rapa,B. oleracea and B. napu
| 保守结构域名称 Motif name | 保守结构域序列 Motif sequence | e值 e-value | 宽度 Width |
|---|---|---|---|
| Motif 1 | C[KN]T[KQ]M[KR]KRN[IV][PT][CF]S[SY]T[TQ]PAQK[IN][DE]V[FL]KPRPR[SL] FTVNNGCSH[LIF][NH]G[ML]P[EK][VA]D[VI][VI]P | 1.4e-672 | 50 |
| Motif 2 | [YS]ELV[SN]NLMDG[EQ]N[MR]WW[EK]SLL[ED]ES[QK][DEQ]P[AD][AG][LI][VF] P[EK][AGS]T[AE]T[KE]K[GL]AT[SF]AFDVEQLW[SN] | 3.1e-601 | 50 |
| Motif 3 | AWTAEED[SN]LLR[QR]CIDKYGEGKWHQVPLRAGL | 8.7e-512 | 31 |
| Motif 4 | K[LF][SN]SDEVDLLLRLHKLLGNRWSLIAGRLPGRTAND[VI]KNYWNTHLSKKHEP | 7.6e-799 | 50 |
| Motif 5 | RCRKSCRLRWLNYLKPSI[KN][RK]G | 3.0e-340 | 21 |
| Motif 6 | [ND][TIA]NNV[CS]EN[SI][IFM]T[CY][NK][KN]D | 2.0e-156 | 15 |
| Motif 7 | ME[GD]S[SP]KGL[RKT]KG | 1.2e-113 | 11 |
| Motif 8 | [LM]LDGET[VG][ET] | 7.2e-62 | 8 |
| Motif 9 | [LP][CF]LG[LFV]N | 1.4e-20 | 6 |
表3 拟南芥、白菜、甘蓝和甘蓝型油菜PAP1/2同源基因保守结构域基序
Table 3 Conserved motif sequence of PAP1/2 homologous genes in Arabidopsis,Brassica rapa,B. oleracea and B. napus
| 保守结构域名称 Motif name | 保守结构域序列 Motif sequence | e值 e-value | 宽度 Width |
|---|---|---|---|
| Motif 1 | C[KN]T[KQ]M[KR]KRN[IV][PT][CF]S[SY]T[TQ]PAQK[IN][DE]V[FL]KPRPR[SL] FTVNNGCSH[LIF][NH]G[ML]P[EK][VA]D[VI][VI]P | 1.4e-672 | 50 |
| Motif 2 | [YS]ELV[SN]NLMDG[EQ]N[MR]WW[EK]SLL[ED]ES[QK][DEQ]P[AD][AG][LI][VF] P[EK][AGS]T[AE]T[KE]K[GL]AT[SF]AFDVEQLW[SN] | 3.1e-601 | 50 |
| Motif 3 | AWTAEED[SN]LLR[QR]CIDKYGEGKWHQVPLRAGL | 8.7e-512 | 31 |
| Motif 4 | K[LF][SN]SDEVDLLLRLHKLLGNRWSLIAGRLPGRTAND[VI]KNYWNTHLSKKHEP | 7.6e-799 | 50 |
| Motif 5 | RCRKSCRLRWLNYLKPSI[KN][RK]G | 3.0e-340 | 21 |
| Motif 6 | [ND][TIA]NNV[CS]EN[SI][IFM]T[CY][NK][KN]D | 2.0e-156 | 15 |
| Motif 7 | ME[GD]S[SP]KGL[RKT]KG | 1.2e-113 | 11 |
| Motif 8 | [LM]LDGET[VG][ET] | 7.2e-62 | 8 |
| Motif 9 | [LP][CF]LG[LFV]N | 1.4e-20 | 6 |
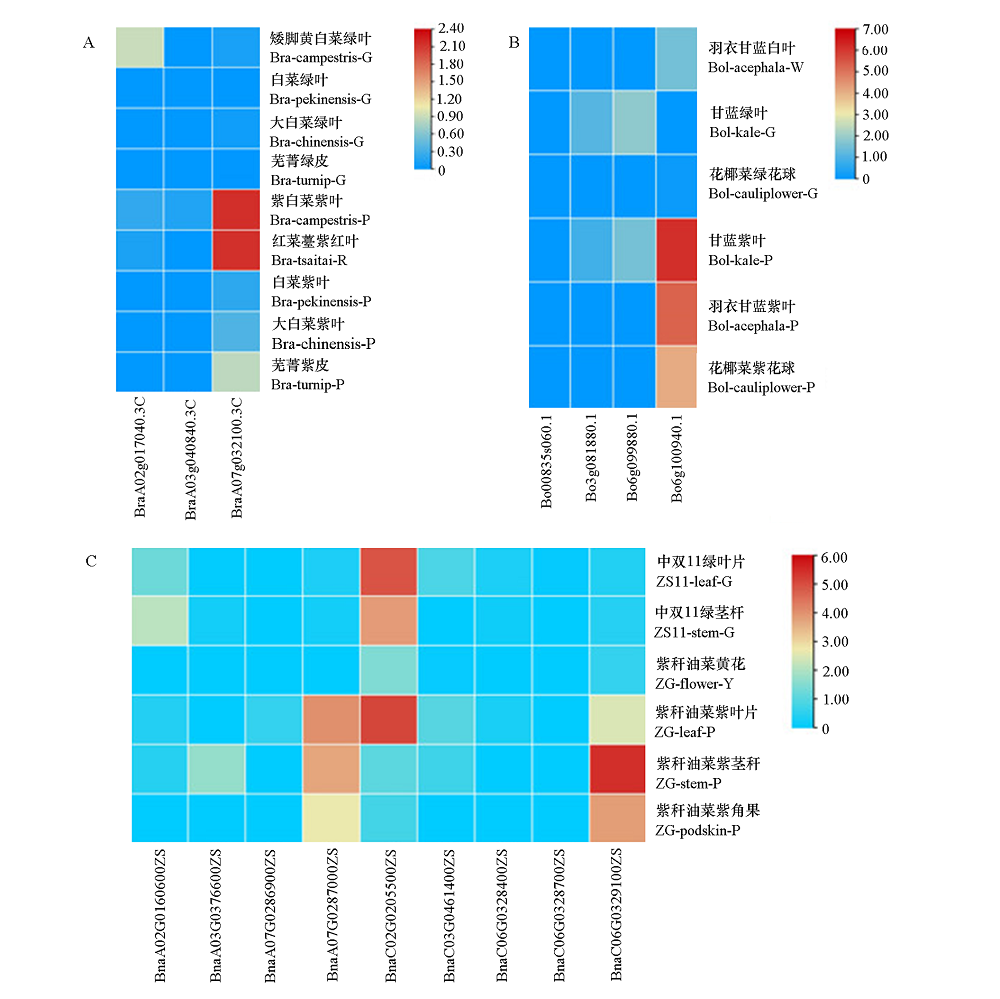
图5 白菜(A)、甘蓝(B)和甘蓝型油菜(C)不同组织PAP1/2同源基因表达热图
Fig. 5 The expression heat map of PAP1/2 homologous genes in different tissues of Brassica rapa(A),B. oleracea(B)and B. napus(C)
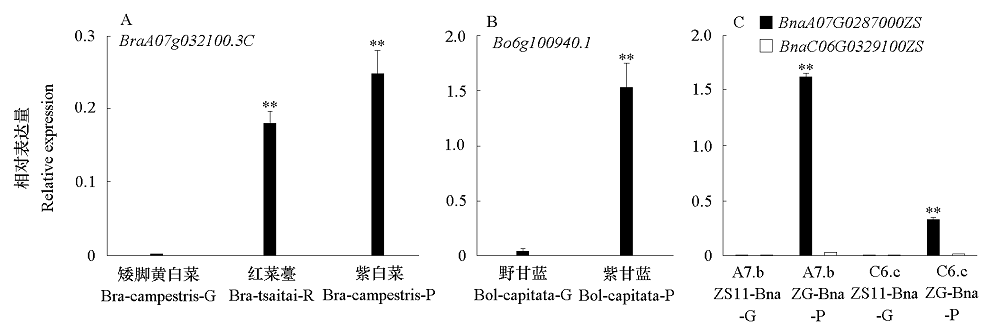
图7 白菜(A)、甘蓝(B)和甘蓝型油菜(B)叶片PAP1/2同源基因qRT-PCR验证
Fig. 7 qRT-PCR analysis of PAP1/2 homologous genes in Brassica rapa(A),B. oleracea(B)and B. napus(C)of leaf
| [1] |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic:a flexible trimmer for illumina sequence data. Bioinformatics, 30 (15):2114-2120.
doi: 10.1093/bioinformatics/btu170 URL |
| [2] |
Borevitz J O, Xia Y, Blount J, Dixon R A, Lamb C. 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell, 12 (12):2383-2394.
pmid: 11148285 |
| [3] |
Chalhoub B, Denoeud F, Liu S, Parkin I A, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh C S, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger P P, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier M C, Fan G, Renault V, Bayer P E, Golicz A A, Manoli S, Lee T H, Thi V H, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom C H, Wang X, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu Z, Sun F, Lim Y P, Lyons E, Town C D, Bancroft I, Wang X, Meng J, Ma J, Pires J C, King G J, Brunel D, Delourme R, Renard M, Aury J M, Adams K L, Batley J, Snowdon R J, Tost J, Edwards D, Zhou Y, Hua W, Sharpe A G, Paterson A H, Guan C, Wincker P. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science, 345 (6199):950-953.
doi: 10.1126/science.1253435 pmid: 25146293 |
| [4] |
Chen D Z, Liu Y, Yin S, Qiu J, Jin Q D, King G J, Wang J, Ge X H, Li Z Y. 2020. Identification and functional analysis of BnaPAP2.A7 revealed that its alternative splicing isoforms play opposite roles in anthocyanin biosynthesis in Brassica napus L. Front Plant Sci, 11:983.
doi: 10.3389/fpls.2020.00983 URL |
| [5] | Cheng F, Wu J, Fang L, Wang X. 2012. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci, 30:198. |
| [6] | Cheng Y, Zhu W, Chen Y, Ito S, Asami T, Wang X. 2014. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD 40 complex by GSK3-like kinases. Elife, 25:e02525. |
| [7] |
Cone K C, Burr F A, Burr B. 1986. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA, 83:9631-9635.
doi: 10.1073/pnas.83.24.9631 URL |
| [8] |
Cone K C, Cocciolone S M, Burr F A, Burr B. 1993. Maize anthocyanin regulatory gene pl is a duplicate of c 1 that functions in the plant. Plant Cell, 5 (12):1795-1805.
pmid: 8305872 |
| [9] |
Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M. 1999. The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J, 19 (4):387-398.
pmid: 10504561 |
| [10] |
Fu Z Z, Shang H Q, Jiang H, Gao J, Dong X Y, Wang H J, Li Y M, Wang L M, Zhang J, Shu Q Y, Chao Y C, Xu M L, Wang R, Wang L S, Zhang H C. 2020. Systematic identification of the light-quality responding anthocyanin synthesis-related transcripts in petunia petals. Horticultural Plant Journal, 6 (6):428-438.
doi: 10.1016/j.hpj.2020.11.006 URL |
| [11] |
Guo N, Cheng F, Wu J, Liu B, Zheng S, Liang J, Wang X. 2014. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics, 15:426.
doi: 10.1186/1471-2164-15-426 URL |
| [12] |
He Q, Wu J, Xue Y, Zhao W, Li R, Zhang L. 2020. The novel gene BrMYB2,located on chromosome A07,with a short intron 1 controls the purple-head trait of Chinese cabbage(Brassica rapa L.). Hortic Res, 7:97.
doi: 10.1038/s41438-020-0319-z URL |
| [13] |
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S L. 2013. TopHat2:accurate alignment of transcriptomes in the presence of insertions,deletions and gene fusions. Genome Biol, 14 (4):R36.
doi: 10.1186/gb-2013-14-4-r36 URL |
| [14] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X:molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 35 (6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [15] | Li Chonghui, Yang Guangsui, Zhang Zhiqun, Yin Junmei. 2021. A novel R2R3-MYB transcription factor gene AaMYB6 involved in anthocyanin biosynthesis in Anthurium andraeanum. Acta Horticulturae Sinica, 48 (10):1859-1872. (in Chinese) |
| 李崇晖, 杨光穗, 张志群, 尹俊梅. 2021. 红掌R2R3-MYB转录因子基因AaMYB6调控花青素苷合成. 园艺学报, 48 (10):1859-1872. | |
| [16] |
Li Y M, Zeng X Z, Guo H, Luo Y W, Kear P, Zhang S M, Zhu G T. 2021. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Horticultural Plant Journal, 7 (2):129-141.
doi: 10.1016/j.hpj.2020.12.001 URL |
| [17] |
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin I A, Zhao M, Ma J, Yu J, Huang S, Wang X, Wang J, Lu K, Fang Z, Bancroft I, Yang T J, Hu Q, Wang X, Yue Z, Li H, Yang L, Wu J, Zhou Q, Wang W, King G J, Pires J C, Lu C, Wu Z, Sampath P, Wang Z, Guo H, Pan S, Yang L, Min J, Zhang D, Jin D, Li W, Belcram H, Tu J, Guan M, Qi C, Du D, Li J, Jiang L, Batley J, Sharpe A G, Park B S, Ruperao P, Cheng F, Waminal N E, Huang Y, Dong C, Wang L, Li J, Hu Z, Zhuang M, Huang Y, Huang J, Shi J, Mei D, Liu J, Lee T H, Wang J, Jin H, Li Z, Li X, Zhang J, Xiao L, Zhou Y, Liu Z, Liu X, Qin R, Tang X, Liu W, Wang Y, Zhang Y, Lee J, Kim H H, Denoeud F, Xu X, Liang X, Hua W, Wang X, Wang J, Chalhoub B, Paterson A H. 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun, 5:3930.
doi: 10.1038/ncomms4930 URL |
| [18] | Liu Xin, Chen Yunzhu, Kim Pyol, Kim Min-Jun, Song Hyondok, Li Yuhua, Wang Yu. 2020. Progress on molecular mechanism and regulation of tomato fruit color formation. Acta Horticulturae Sinica, 47 (9):1689-1704. (in Chinese) |
| 刘昕, 陈韵竹, Kim Pyol, Kim Min-Jun, Song Hyondok, 李玉花, 王宇. 2020. 番茄果实颜色形成的分子机制及调控研究进展. 园艺学报, 47 (9):1689-1704. | |
| [19] |
Meng J X, Gao Y, Han M L, Liu P Y, Yang C, Shen T, Li H H. 2020. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Horticultural Plant Journal, 6 (5):284-292.
doi: 10.1016/j.hpj.2020.06.004 URL |
| [20] |
Pertea M, Kim D, Pertea G M, Leek J T, Salzberg S L. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT,StringTie and Ballgown. Nat Protoc, 11 (9):1650-1667.
doi: 10.1038/nprot.2016.095 URL |
| [21] |
Shi M Z, Xie D Y. 2014. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotechnol, 8 (1):47-60.
doi: 10.2174/1872208307666131218123538 URL |
| [22] |
Tanaka M, Fujimori T, Uchida I, Yamaguchi S, Takeda K. 2001. A malonylated,anthocyanin and flavonols in blue Meconopsis flowers. Phytochemistry, 56 (4):373-386.
pmid: 11249104 |
| [23] |
Tanaka Y, Brugliera F, Chandler S. 2009. Recent progress of flower colour modification by biotechnology. Int J Mol Sci, 10 (12):5350-5369.
doi: 10.3390/ijms10125350 URL |
| [24] |
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J H, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires J C, Paterson A H, Chalhoub B, Wang B, Hayward A, Sharpe A G, Park B S, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King G J, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin I A, Batley J, Kim J S, Just J, Li J, Xu J, Deng J, Kim J A, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links M G, Zhao M, Jin M, Ramchiary N, Drou N, Berkman P J, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon S J, Choi S R, Lee T H, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z. 2011. Brassica rapa genome sequencing project consortium. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet, 43 (10):1035-1039.
doi: 10.1038/ng.919 URL |
| [25] |
Xi H C, He Y J, Chen H Y. 2021. Functional characterization of SmbHLH13in anthocyanin biosynthesis and flowering in eggplant. Horticultural Plant Journal, 7 (1):73-80.
doi: 10.1016/j.hpj.2020.08.006 URL |
| [26] |
Xu W, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci, 20 (3):176-185.
doi: 10.1016/j.tplants.2014.12.001 URL |
| [27] |
Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C. 2014. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol, 202 (1):132-144.
doi: 10.1111/nph.12620 URL |
| [28] |
Yan C, An G, Zhu T, Zhang W, Zhang L, Peng L, Chen J, Kuang H. 2019. Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor Appl Genet, 132 (4):895-906.
doi: 10.1007/s00122-018-3245-9 URL |
| [29] |
Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, Hu Z, Chen S, Pental D, Ju Y, Yao P, Li X, Xie K, Zhang J, Wang J, Liu F, Ma W, Shopan J, Zheng H, Mackenzie S A, Zhang M. 2016. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet, 48 (10):1225-1232.
doi: 10.1038/ng.3657 URL |
| [30] |
Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T. 2013. The R2R3-MYB,bHLH,WD40,and related transcription factors in flavonoid biosynthesis. Funct Integr Genomics, 13 (1):75-98.
doi: 10.1007/s10142-012-0301-4 URL |
| [31] |
Zhou L L, Shi M Z, Xie D Y. 2012. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana. Planta, 236 (3):825-837.
doi: 10.1007/s00425-012-1674-2 URL |
| [1] | 叶子茂, 申晚霞, 刘梦雨, 王 彤, 张晓楠, 余 歆, 刘小丰, 赵晓春, . R2R3-MYB转录因子CitMYB21对柑橘类黄酮生物合成的影响[J]. 园艺学报, 2023, 50(2): 250-264. |
| [2] | 韩 睿, 钟雄辉, 陈登辉, 崔 建, 乐祥庆, 颉建明, 康俊根, . 黑腐病菌效应因子XopR的甘蓝靶标基因BobHLH34的克隆及功能分析[J]. 园艺学报, 2023, 50(2): 319-330. |
| [3] | 崔建, 钟雄辉, 刘泽慈, 陈登辉, 李海龙, 韩睿, 乐祥庆, 康俊根, 王超. 结球甘蓝染色体片段替换系构建[J]. 园艺学报, 2023, 50(1): 65-78. |
| [4] | 韩书辉, 韩彩锋, 韩书荣, 韩彩梅, 韩 旭. 秋大白菜新品种‘胶研秋宝’[J]. 园艺学报, 2022, 49(S2): 83-84. |
| [5] | 汪维红, 张凤兰, 余阳俊, 张德双, 赵岫云, 于拴仓, 苏同兵, 李佩荣, 辛晓云. 秋大白菜新品种‘京秋1518’[J]. 园艺学报, 2022, 49(S2): 85-86. |
| [6] | 余阳俊, 汪维红, 苏同兵, 张凤兰, 张德双, 赵岫云, 于拴仓, 李佩荣, 辛晓云, 王 姣. 抗根肿病耐抽薹大白菜新品种‘京春CR3’[J]. 园艺学报, 2022, 49(S2): 87-88. |
| [7] | 王丽丽, 王 鑫, 吴海东, 温 蔷, 杨晓飞. 抗根肿病大白菜新品种‘辽白28’[J]. 园艺学报, 2022, 49(S2): 89-90. |
| [8] | 余阳俊, 苏同兵, 张凤兰, 张德双, 赵岫云, 于拴仓, 汪维红, 李佩荣, 辛晓云, 王 姣, 武长见. 紫色苗用型大白菜新品种‘京研紫快菜’[J]. 园艺学报, 2022, 49(S2): 91-92. |
| [9] | 黄 鹂, 陈财志, 余小林, 姚祥坦, 曹家树, . 早中熟普通白菜新品种‘浙大青’[J]. 园艺学报, 2022, 49(S2): 93-94. |
| [10] | 吕红豪, 杨丽梅, 方智远, 张扬勇, 庄 木, 刘玉梅, 王 勇, 季家磊, 李占省, 韩风庆. 春甘蓝新品种‘中甘27’和‘中甘28’[J]. 园艺学报, 2022, 49(S1): 63-64. |
| [11] | 周 娜, 陶伟林, 陆景伟, 郑 阳, 胡 燕, 潘晓雪. 甘蓝新品种‘渝园春印’[J]. 园艺学报, 2022, 49(S1): 65-66. |
| [12] | 徐立功, 韩太利, 孙继峰, 杨晓东, 谭金霞. 苗用大白菜新品种‘锦绿2号’[J]. 园艺学报, 2022, 49(S1): 67-68. |
| [13] | 邵贵荣, 朱 彬, 林 晓, 曹 萍, 方 勇, 崔 田, 蒋 鹏, 林咏铭, 林 魁, 林志滔. 白菜新品种‘金品008’[J]. 园艺学报, 2022, 49(S1): 69-70. |
| [14] | 姜悦悦, 王田田, 赵 阳, 汪承刚, 侯金锋, 袁凌云. 白菜新品种‘皖绿2号’[J]. 园艺学报, 2022, 49(S1): 71-72. |
| [15] | 邹 雪, 丁 凡, 刘丽芳, 余韩开宗, 陈年伟, 饶莉萍. 紫色马铃薯新品种‘绵紫芋1号’[J]. 园艺学报, 2022, 49(S1): 93-94. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司