
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (1): 190-202.doi: 10.16420/j.issn.0513-353x.2022-1197
• Plant Protection • Previous Articles Next Articles
CHEN Ruhao1, MENG Fanye1, YANG Manhua1, CHEN Jiaru1, ZOU Ying2, SONG Botao1, MA Hui3, NIE Bihua1,*( )
)
Received:2023-03-03
Revised:2023-10-23
Online:2024-01-25
Published:2024-01-16
Contact:
(E-mail:CHEN Ruhao, MENG Fanye, YANG Manhua, CHEN Jiaru, ZOU Ying, SONG Botao, MA Hui, NIE Bihua. Analysis and Identification of Endogenous Viral Elements in Solanaceous Crop Genome[J]. Acta Horticulturae Sinica, 2024, 51(1): 190-202.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2022-1197
| 引物名称 Primer name | 引物序列 Primer sequences | 大小/bp Size | 引物类型 Primer type | 特异性前引物 + 特异性后引物 Forward + reverse |
|---|---|---|---|---|
| TVCV-1F | CACACTGGTGGTTCACAACACCATG | 396 | 基因内引物 Inner primers | TVCV + TVCV |
| TVCV-1R | GGGCTTGTATGCTTACTATTGC | |||
| TVCV-2F | CACCAATGATAGATACGGGAGC | 506 | 基因内引物 Inner primers | TVCV + TVCV |
| TVCV-2R | CCCATCCTTGTAAAGGATCTTCAC | |||
| TVCV-3F | CACCAATGATAGATACGGGAGC | 294 | 基因内引物 Inner primers | TVCV + TVCV |
| TVCV-3R | CATGGTGTTGTGAACCACCAGTGTG | |||
| TVCV-UF | ATGCTCTTGTCGTAGGTCG | 1 014 | 跨基因引物 Gene-spanning primers | TVCV上游基因 + TVCV Gene-specific primers upstream of TVCV + TVCV |
| TVCV-UR | GTGGACAGTATGGACAATGAGATAC | |||
| TVCV-DF | AGTGGATAGTATGGACAACG | 716 | 跨基因引物 Gene-spanning primers | TVCV + TVCV下游基因 Gene-specific primers upstream of TVCV + TVCV specific primers |
| TVCV-DR | TCTTTACAACTTGTATCGGAGG |
Table 1 The primers for molecular verification
| 引物名称 Primer name | 引物序列 Primer sequences | 大小/bp Size | 引物类型 Primer type | 特异性前引物 + 特异性后引物 Forward + reverse |
|---|---|---|---|---|
| TVCV-1F | CACACTGGTGGTTCACAACACCATG | 396 | 基因内引物 Inner primers | TVCV + TVCV |
| TVCV-1R | GGGCTTGTATGCTTACTATTGC | |||
| TVCV-2F | CACCAATGATAGATACGGGAGC | 506 | 基因内引物 Inner primers | TVCV + TVCV |
| TVCV-2R | CCCATCCTTGTAAAGGATCTTCAC | |||
| TVCV-3F | CACCAATGATAGATACGGGAGC | 294 | 基因内引物 Inner primers | TVCV + TVCV |
| TVCV-3R | CATGGTGTTGTGAACCACCAGTGTG | |||
| TVCV-UF | ATGCTCTTGTCGTAGGTCG | 1 014 | 跨基因引物 Gene-spanning primers | TVCV上游基因 + TVCV Gene-specific primers upstream of TVCV + TVCV |
| TVCV-UR | GTGGACAGTATGGACAATGAGATAC | |||
| TVCV-DF | AGTGGATAGTATGGACAACG | 716 | 跨基因引物 Gene-spanning primers | TVCV + TVCV下游基因 Gene-specific primers upstream of TVCV + TVCV specific primers |
| TVCV-DR | TCTTTACAACTTGTATCGGAGG |
| 病毒名称 Virus name | 病毒ID Virus ID | 科 Family | 属 Genus | 检出率/ Detec- tion rate | 总数 Total number | 总长/bp Total length | 平均长/bp Average length | 检出基因组 Genome containing the virus |
|---|---|---|---|---|---|---|---|---|
| 烟草脉明病毒 Tobacco vein clearing virus | NC_003378.1 | 花椰菜病毒科 Caulimoviridae | 木薯脉花叶病毒属Cavemovirus | 94.3 (33/35) | 45 733 | 57 538 756 | 1 258 | 几乎所有基因组 Almost all genomes |
| 花生褪绿条纹病毒 Peanut chlorotic streak virus | NC_001634.1 | 花椰菜病毒科 Caulimoviridae | 大豆褪绿斑驳病毒属Soymovirus | 2.8 (1/35) | 5 | 4 220 | 844 | 本氏烟草 Nicotiana benthamiana |
| 金黄球藻病毒 Chrysochromulina ericina virus | NC_028094.1 | 藻类DNA病毒科 Phycodnaviridae | 未分类 Unclassified | 48.6 (17/35) | 48 | 35 546 | 741 | 烟草、茄子、矮牵牛 Tobacco,eggplant,petunia |
| 矮牵牛清脉病毒 Petunia vein clearing virus | NC_001839.2 | 花椰菜病毒科 Caulimoviridae | 碧冬茄病毒属 Petuvirus | 40.0 (14/35) | 892 | 550 159 | 617 | 烟草、矮牵牛 Tobacco,petunia |
| 木薯脉花叶病毒 Cassava vein mosaic virus | NC_001648.1 | 花椰菜病毒科 Caulimoviridae | 木薯脉花叶病毒属Cavemovirus | 28.6 (10/35) | 63 | 22 235 | 353 | 马铃薯、番茄、辣椒、茄子Potato,tomato,pepper,eggplant |
| 甘薯清脉病毒 Sweet potato vein clearing virus | NC_015228.1 | 花椰菜病毒科 Caulimoviridae | 索伦病毒属Solendovirus | 25.7 (9/35) | 112 | 26 347 | 235 | 番茄、烟草 Tomato,tobacco |
| 棉花曲叶木尔坦病毒 Cotton leaf curl Multan virus | NC_007721.1 | 双生病毒科 Geminiviridae | 菜豆金色花叶病毒Begomovirus | 22.9 (8/35) | 1 590 | 351 770 | 221 | 烟草 Tobacco |
| 尤卡坦黄斑病毒 Capraria yellow spot Yucatan virus | NC_022007.1 | 双生病毒科 Geminiviridae | 菜豆金色花叶病毒Begomovirus | 11.4 (4/35) | 20 | 4 308 | 215 | 普通烟草 Nicotiana tabacum |
| 甘薯茎状病毒 Sweet potato caulimo- Like virus | NC_015328.1 | 花椰菜病毒科 Caulimoviridae | 木薯脉花叶病毒属Cavemovirus | 14.3 (5/35) | 113 | 20 335 | 180 | 茄子、辣椒、马铃薯 Eggplant,pepper,potato |
| 棉皱叶病毒 Cotton leaf crumple virus | NC_004580.1 | 双生病毒科 Geminiviridae | 菜豆金色花叶病毒Begomovirus | 11.4 (4/35) | 34 | 4 700 | 138 | 普通烟草 Nicotiana tabacum |
| 马铃薯卷叶病毒 Potato leafroll virus | NC_001747.1 | 黄症病毒科 Luteoviridae | 马铃薯卷叶病毒属Polerovirus | 48.6 (17/35) | 155 | 16 283 | 105 | 辣椒、烟草、矮牵牛 Pepper,tobacco, petunia |
| 香蕉条纹ca病毒 Banana streak ca Virus | NC_015506.1 | 花椰菜病毒科 Caulimoviridae | 杆状DNA病毒属Badnavirus | 11.4 (4/35) | 18 | 831 | 46 | 辣椒、矮牵牛 Pepper,petunia |
| 柑橘内源副逆转录病毒citrus endogenous Pararetrovirus | NC_023153.1 | 逆转录病毒科 Retroviridae | 未知 Unknown | 54.3 (19/35) | 444 | 19 616 | 44 | 烟草、矮牵牛、番茄 Tobacco,petunia, Tomato |
| 大丽花花叶病毒 Dahlia mosaic virus | NC_018616.1 | 花椰菜病毒科 Caulimoviridae | 花椰菜花叶病毒属Caulimovirus | 22.9 (8/35) | 45 | 1 799 | 40 | 辣椒、矮牵牛、马铃薯Pepper,petunia, potato |
| 香蕉条纹糠秕病毒 Banana streak Mysore virus | NC_006955.1 | 花椰菜病毒科 Caulimoviridae | 杆状DNA病毒属Badnavirus | 2.8 (1/35) | 4 | 139 | 35 | 马铃薯品种 Solanum tuberosum commersonii |
| 天竺葵脉带病毒 Pelargonium vein Banding virus | NC_013262.1 | 花椰菜病毒科 Caulimoviridae | 杆状DNA病毒属Badnavirus | 5.7 (2/35) | 4 | 144 | 36 | 醋栗番茄 Solanum pimpinellifolium |
| 水稻东格鲁杆状病毒 Rice tungro bacilliform virus | NC_001914.1 | 花椰菜病毒科 Caulimoviridae | 东格鲁病毒属 Tungrovirus | 11.4 (4/35) | 15 | 495 | 33 | 烟草Tobacco |
Table 2 The plant EVEs in the genome of six Solanaceae crops
| 病毒名称 Virus name | 病毒ID Virus ID | 科 Family | 属 Genus | 检出率/ Detec- tion rate | 总数 Total number | 总长/bp Total length | 平均长/bp Average length | 检出基因组 Genome containing the virus |
|---|---|---|---|---|---|---|---|---|
| 烟草脉明病毒 Tobacco vein clearing virus | NC_003378.1 | 花椰菜病毒科 Caulimoviridae | 木薯脉花叶病毒属Cavemovirus | 94.3 (33/35) | 45 733 | 57 538 756 | 1 258 | 几乎所有基因组 Almost all genomes |
| 花生褪绿条纹病毒 Peanut chlorotic streak virus | NC_001634.1 | 花椰菜病毒科 Caulimoviridae | 大豆褪绿斑驳病毒属Soymovirus | 2.8 (1/35) | 5 | 4 220 | 844 | 本氏烟草 Nicotiana benthamiana |
| 金黄球藻病毒 Chrysochromulina ericina virus | NC_028094.1 | 藻类DNA病毒科 Phycodnaviridae | 未分类 Unclassified | 48.6 (17/35) | 48 | 35 546 | 741 | 烟草、茄子、矮牵牛 Tobacco,eggplant,petunia |
| 矮牵牛清脉病毒 Petunia vein clearing virus | NC_001839.2 | 花椰菜病毒科 Caulimoviridae | 碧冬茄病毒属 Petuvirus | 40.0 (14/35) | 892 | 550 159 | 617 | 烟草、矮牵牛 Tobacco,petunia |
| 木薯脉花叶病毒 Cassava vein mosaic virus | NC_001648.1 | 花椰菜病毒科 Caulimoviridae | 木薯脉花叶病毒属Cavemovirus | 28.6 (10/35) | 63 | 22 235 | 353 | 马铃薯、番茄、辣椒、茄子Potato,tomato,pepper,eggplant |
| 甘薯清脉病毒 Sweet potato vein clearing virus | NC_015228.1 | 花椰菜病毒科 Caulimoviridae | 索伦病毒属Solendovirus | 25.7 (9/35) | 112 | 26 347 | 235 | 番茄、烟草 Tomato,tobacco |
| 棉花曲叶木尔坦病毒 Cotton leaf curl Multan virus | NC_007721.1 | 双生病毒科 Geminiviridae | 菜豆金色花叶病毒Begomovirus | 22.9 (8/35) | 1 590 | 351 770 | 221 | 烟草 Tobacco |
| 尤卡坦黄斑病毒 Capraria yellow spot Yucatan virus | NC_022007.1 | 双生病毒科 Geminiviridae | 菜豆金色花叶病毒Begomovirus | 11.4 (4/35) | 20 | 4 308 | 215 | 普通烟草 Nicotiana tabacum |
| 甘薯茎状病毒 Sweet potato caulimo- Like virus | NC_015328.1 | 花椰菜病毒科 Caulimoviridae | 木薯脉花叶病毒属Cavemovirus | 14.3 (5/35) | 113 | 20 335 | 180 | 茄子、辣椒、马铃薯 Eggplant,pepper,potato |
| 棉皱叶病毒 Cotton leaf crumple virus | NC_004580.1 | 双生病毒科 Geminiviridae | 菜豆金色花叶病毒Begomovirus | 11.4 (4/35) | 34 | 4 700 | 138 | 普通烟草 Nicotiana tabacum |
| 马铃薯卷叶病毒 Potato leafroll virus | NC_001747.1 | 黄症病毒科 Luteoviridae | 马铃薯卷叶病毒属Polerovirus | 48.6 (17/35) | 155 | 16 283 | 105 | 辣椒、烟草、矮牵牛 Pepper,tobacco, petunia |
| 香蕉条纹ca病毒 Banana streak ca Virus | NC_015506.1 | 花椰菜病毒科 Caulimoviridae | 杆状DNA病毒属Badnavirus | 11.4 (4/35) | 18 | 831 | 46 | 辣椒、矮牵牛 Pepper,petunia |
| 柑橘内源副逆转录病毒citrus endogenous Pararetrovirus | NC_023153.1 | 逆转录病毒科 Retroviridae | 未知 Unknown | 54.3 (19/35) | 444 | 19 616 | 44 | 烟草、矮牵牛、番茄 Tobacco,petunia, Tomato |
| 大丽花花叶病毒 Dahlia mosaic virus | NC_018616.1 | 花椰菜病毒科 Caulimoviridae | 花椰菜花叶病毒属Caulimovirus | 22.9 (8/35) | 45 | 1 799 | 40 | 辣椒、矮牵牛、马铃薯Pepper,petunia, potato |
| 香蕉条纹糠秕病毒 Banana streak Mysore virus | NC_006955.1 | 花椰菜病毒科 Caulimoviridae | 杆状DNA病毒属Badnavirus | 2.8 (1/35) | 4 | 139 | 35 | 马铃薯品种 Solanum tuberosum commersonii |
| 天竺葵脉带病毒 Pelargonium vein Banding virus | NC_013262.1 | 花椰菜病毒科 Caulimoviridae | 杆状DNA病毒属Badnavirus | 5.7 (2/35) | 4 | 144 | 36 | 醋栗番茄 Solanum pimpinellifolium |
| 水稻东格鲁杆状病毒 Rice tungro bacilliform virus | NC_001914.1 | 花椰菜病毒科 Caulimoviridae | 东格鲁病毒属 Tungrovirus | 11.4 (4/35) | 15 | 495 | 33 | 烟草Tobacco |
| 物种 Species | 植物基因组大小/bp Assembly size of plant genome | 内源性TVCV Endogenous TVCV | 数据库 Data- base | |||
|---|---|---|---|---|---|---|
| 覆盖大小Coverage size | 占植物基因组百分比 Percentagein plant genome | 数量 Number | 平均长度/bp The average length | |||
| Capsicum annuum‘CM334’ | 2 753 501 891 | 8 829 122 | 0.320651 | 5 341 | 1 653 | SOL |
| 3 106 362 892 | 9 863 964 | 0.317541 | 6 379 | 1 546 | NCBI | |
| Capsicum annuum‘Zunla-1’ | 3 420 028 601 | 6 640 658 | 0.194170 | 4 580 | 1 450 | SOL |
| 2 972 093 730 | 7 472 302 | 0.251415 | 5 139 | 1 454 | NCBI | |
| Capsicum annuum var. glabriusculum | 3 586 841 310 | 3 990 309 | 0.111249 | 3 080 | 1 296 | SOL |
| 2 803 123 110 | 4 678 985 | 0.166920 | 3 575 | 1 309 | NCBI | |
| Nicotiana benthamiana | 3 020 841 706 | 4 409 | 0.000146 | 5 | 882 | SOL |
| 76 127 810 | 0 | 0 | 0 | 0 | NCBI | |
| Nicotiana otophora | 2 749 613 483 | 278 606 | 0.010133 | 756 | 369 | SOL |
| 2 805 105 985 | 278 606 | 0.009932 | 756 | 369 | NCBI | |
| Nicotiana sylvestris | 2 258 816 203 | 321 141 | 0.014217 | 564 | 569 | SOL |
| 2 280 242 707 | 321 141 | 0.014084 | 564 | 569 | NCBI | |
| Nicotiana tabacum Basma Xanthi | 3 805 503 023 | 440 123 | 0.011565 | 928 | 474 | SOL |
| 3 854 148 512 | 440 361 | 0.011426 | 931 | 473 | NCBI | |
| Nicotiana tabacum K326 (Flue-cured) | 3 799 828 677 | 463 973 | 0.012210 | 991 | 468 | SOL |
| 3 840 635 360 | 464 426 | 0.012092 | 999 | 465 | NCBI | |
| Nicotiana tabacum TN90 (Burley) | 3 772 618 817 | 624 141 | 0.016544 | 1 062 | 588 | SOL |
| 3 802 293 918 | 624 217 | 0.016417 | 1 063 | 587 | NCBI | |
| Nicotiana tomentosiformis | 1 715 755 740 | 185 908 | 0.010835 | 516 | 360 | SOL |
| 1 730 121 004 | 185 908 | 0.010745 | 516 | 360 | NCBI | |
| Petunia axillaris N | 1 281 752 908 | 0 | 0 | 0 | 0 | SOL |
| Petunia inflate S6 | 1 312 457 388 | 321 | 0.000024 | 6 | 54 | SOL |
| Solanum arcanum LA2157 | 678 871 964 | 548 248 | 0.080759 | 350 | 1 566 | NCBI |
| Solanum habrochaites LYC4 | 738 507 092 | 986 200 | 0.133540 | 554 | 1 780 | NCBI |
| Solanum lycopersicum Heinz 1706 | 835 714 844 | 1 475 086 | 0.176506 | 710 | 2 078 | SOL |
| 834 512 558 | 1 475 086 | 0.176760 | 710 | 2 078 | NCBI | |
| Solanum pennellii LA716 | 1 003 809 056 | 1 299 035 | 0.129411 | 559 | 2 324 | SOL |
| 938 007 464 | 1 238 991 | 0.132088 | 519 | 2 387 | NCBI | |
| Solanum pimpinellifolium LA1589 | 716 421 466 | 274 456 | 0.038309 | 481 | 571 | SOL |
| 728 838 890 | 274 465 | 0.037658 | 481 | 571 | NCBI | |
| Solanum tuberosum DM1-3 | 735 374 925 | 697 015 | 0.094784 | 684 | 1 019 | SOL |
| 716 822 822 | 852 693 | 0.118954 | 840 | 1 015 | NCBI | |
| Solanum commersonii | 744 831 912 | 2 080 946 | 0.279385 | 1 652 | 1 260 | NCBI |
| Solanum melongena | 847 552 547 | 113 957 | 0.013445 | 221 | 516 | SOL |
| 846 607 191 | 113 957 | 0.013460 | 221 | 516 | NCBI | |
Table 3 The contributions of endogenous TVCV to the genomes of various Solanaceae plant species
| 物种 Species | 植物基因组大小/bp Assembly size of plant genome | 内源性TVCV Endogenous TVCV | 数据库 Data- base | |||
|---|---|---|---|---|---|---|
| 覆盖大小Coverage size | 占植物基因组百分比 Percentagein plant genome | 数量 Number | 平均长度/bp The average length | |||
| Capsicum annuum‘CM334’ | 2 753 501 891 | 8 829 122 | 0.320651 | 5 341 | 1 653 | SOL |
| 3 106 362 892 | 9 863 964 | 0.317541 | 6 379 | 1 546 | NCBI | |
| Capsicum annuum‘Zunla-1’ | 3 420 028 601 | 6 640 658 | 0.194170 | 4 580 | 1 450 | SOL |
| 2 972 093 730 | 7 472 302 | 0.251415 | 5 139 | 1 454 | NCBI | |
| Capsicum annuum var. glabriusculum | 3 586 841 310 | 3 990 309 | 0.111249 | 3 080 | 1 296 | SOL |
| 2 803 123 110 | 4 678 985 | 0.166920 | 3 575 | 1 309 | NCBI | |
| Nicotiana benthamiana | 3 020 841 706 | 4 409 | 0.000146 | 5 | 882 | SOL |
| 76 127 810 | 0 | 0 | 0 | 0 | NCBI | |
| Nicotiana otophora | 2 749 613 483 | 278 606 | 0.010133 | 756 | 369 | SOL |
| 2 805 105 985 | 278 606 | 0.009932 | 756 | 369 | NCBI | |
| Nicotiana sylvestris | 2 258 816 203 | 321 141 | 0.014217 | 564 | 569 | SOL |
| 2 280 242 707 | 321 141 | 0.014084 | 564 | 569 | NCBI | |
| Nicotiana tabacum Basma Xanthi | 3 805 503 023 | 440 123 | 0.011565 | 928 | 474 | SOL |
| 3 854 148 512 | 440 361 | 0.011426 | 931 | 473 | NCBI | |
| Nicotiana tabacum K326 (Flue-cured) | 3 799 828 677 | 463 973 | 0.012210 | 991 | 468 | SOL |
| 3 840 635 360 | 464 426 | 0.012092 | 999 | 465 | NCBI | |
| Nicotiana tabacum TN90 (Burley) | 3 772 618 817 | 624 141 | 0.016544 | 1 062 | 588 | SOL |
| 3 802 293 918 | 624 217 | 0.016417 | 1 063 | 587 | NCBI | |
| Nicotiana tomentosiformis | 1 715 755 740 | 185 908 | 0.010835 | 516 | 360 | SOL |
| 1 730 121 004 | 185 908 | 0.010745 | 516 | 360 | NCBI | |
| Petunia axillaris N | 1 281 752 908 | 0 | 0 | 0 | 0 | SOL |
| Petunia inflate S6 | 1 312 457 388 | 321 | 0.000024 | 6 | 54 | SOL |
| Solanum arcanum LA2157 | 678 871 964 | 548 248 | 0.080759 | 350 | 1 566 | NCBI |
| Solanum habrochaites LYC4 | 738 507 092 | 986 200 | 0.133540 | 554 | 1 780 | NCBI |
| Solanum lycopersicum Heinz 1706 | 835 714 844 | 1 475 086 | 0.176506 | 710 | 2 078 | SOL |
| 834 512 558 | 1 475 086 | 0.176760 | 710 | 2 078 | NCBI | |
| Solanum pennellii LA716 | 1 003 809 056 | 1 299 035 | 0.129411 | 559 | 2 324 | SOL |
| 938 007 464 | 1 238 991 | 0.132088 | 519 | 2 387 | NCBI | |
| Solanum pimpinellifolium LA1589 | 716 421 466 | 274 456 | 0.038309 | 481 | 571 | SOL |
| 728 838 890 | 274 465 | 0.037658 | 481 | 571 | NCBI | |
| Solanum tuberosum DM1-3 | 735 374 925 | 697 015 | 0.094784 | 684 | 1 019 | SOL |
| 716 822 822 | 852 693 | 0.118954 | 840 | 1 015 | NCBI | |
| Solanum commersonii | 744 831 912 | 2 080 946 | 0.279385 | 1 652 | 1 260 | NCBI |
| Solanum melongena | 847 552 547 | 113 957 | 0.013445 | 221 | 516 | SOL |
| 846 607 191 | 113 957 | 0.013460 | 221 | 516 | NCBI | |

Fig. 1 The PCR detection result of endogenous TVCV sequences in potato varieties DM1-3,CMM60-2,Shebody,Desiree,40-3 and E3 M:Marker. The DNA and RNA of each variety were sequentially detected using five pairs of(1-5)primers(Table 1).
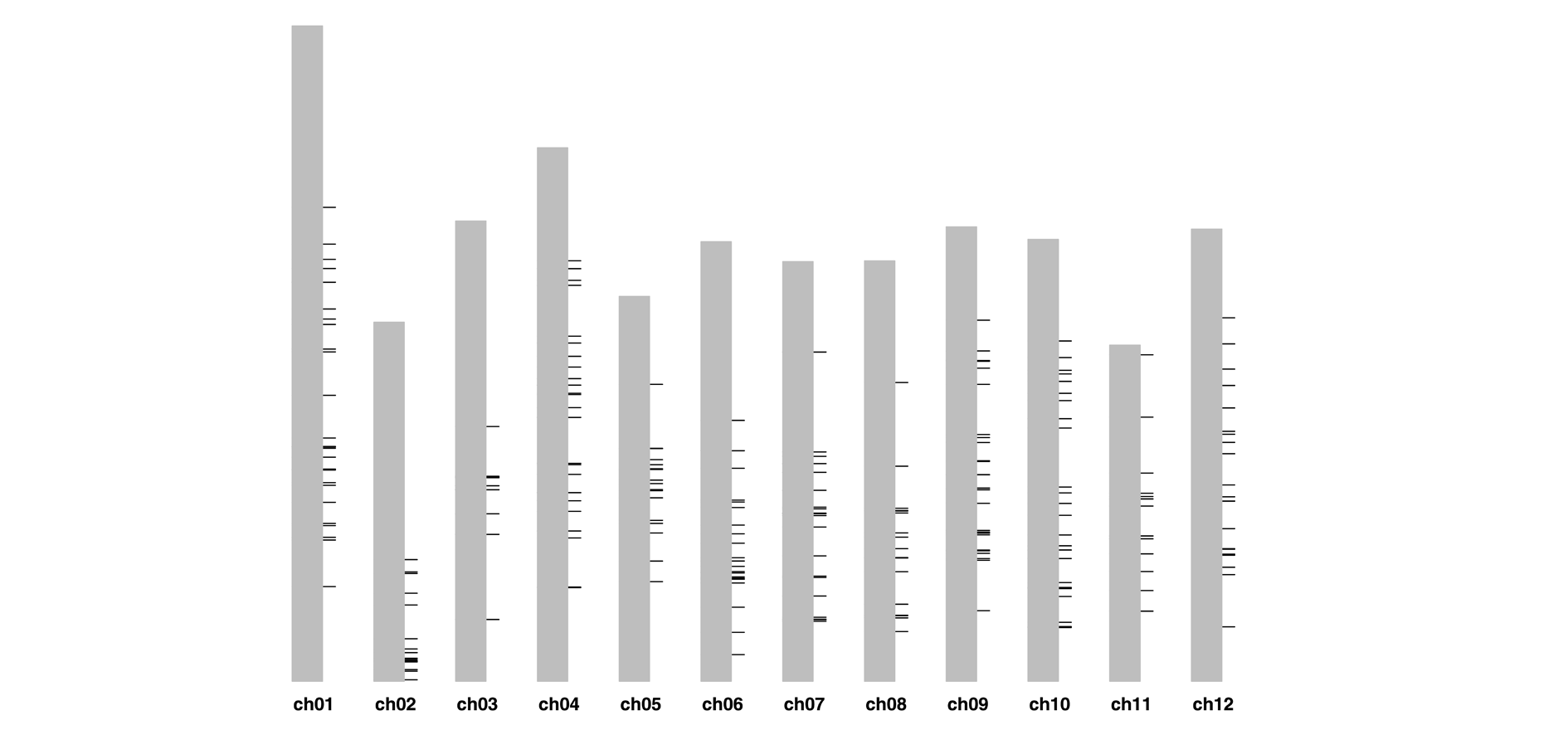
Fig. 2 Position distribution of endogenous TVCV on DM1-3 chromosome The black short line represents the endogenous TVCV sequence,and the thickness of the line represents the number of TVCV sequences.
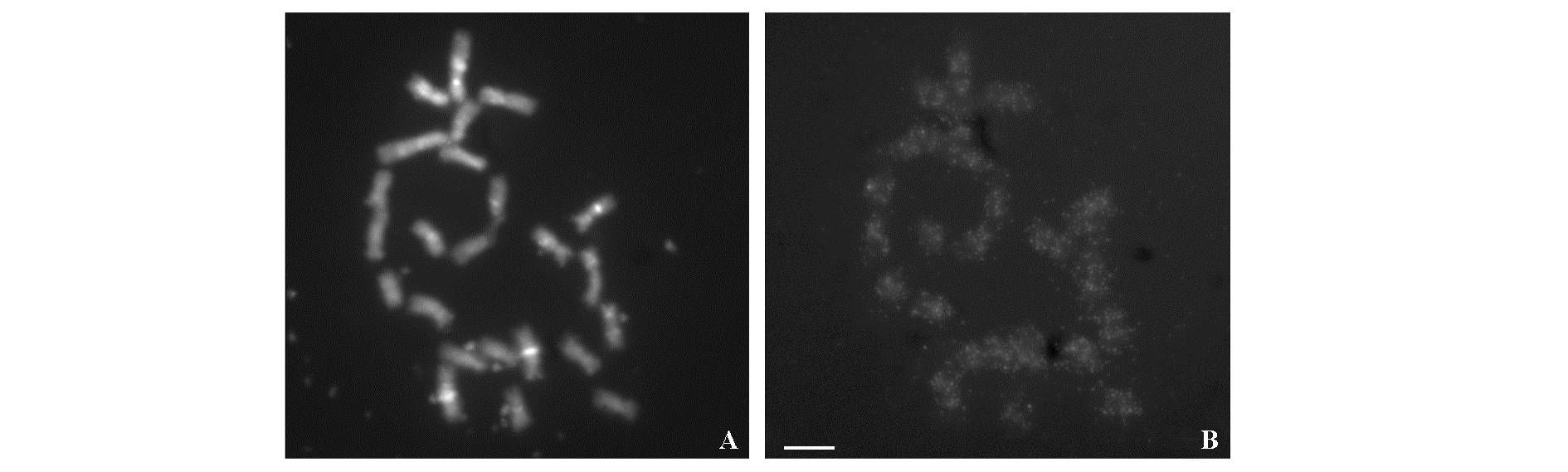
Fig. 3 FISH mapping of endogenous TVCV on DM1-3 chromosome A:Fluorescence signal of DM1-3 24 chromosomes after staining;B:Fluorescence signal of DM1-3 after in situ hybridization with TVCV probes.

Fig. 4 Position distribution of endogenous TVCV on TVCV genome The gray line represents the TVCV genome,the black line represents the coding region of TVCV. The red and blue lines represent the TVCV sequence inserted in the DM1-3 genome in the forward and reverse directions,respectively. The length and coordinate information of the lines represent the sequence length of TVCV inserted in the DM1-3 genome and the position information on the TVCV genome.
| [1] |
doi: 10.1270/jsbbs.53.177 URL |
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
doi: 10.1186/1745-6150-4-21 pmid: 19558678 |
| [6] |
doi: 10.1186/s12985-020-01472-1 |
| [7] |
doi: 10.1128/JVI.00899-13 pmid: 23720724 |
| [8] |
doi: 10.1111/mpp.13019 pmid: 33231927 |
| [9] |
doi: 10.1016/j.coviro.2013.08.012 pmid: 24035682 |
| [10] |
doi: 10.1111/mpp.12490 pmid: 27870389 |
| [11] |
doi: 10.1016/j.cpb.2014.07.002 URL |
| [12] |
doi: 10.1038/s41598-017-16399-x pmid: 29330451 |
| [13] |
doi: 10.1038/463039a |
| [14] |
doi: 10.1038/nrg3199 pmid: 22421730 |
| [15] |
doi: S1879-6257(17)30021-4 pmid: 28818736 |
| [16] |
doi: 10.1128/JVI.00401-10 URL |
| [17] |
doi: 10.1038/ncomms6269 |
| [18] |
doi: 10.1126/science.1167375 pmid: 19150848 |
| [19] |
|
| [20] |
|
| [21] |
doi: 10.1159/000084989 pmid: 16093709 |
| [22] |
|
| [23] |
doi: 10.1038/nature08695 |
| [24] |
|
| [25] |
|
| [26] |
pmid: 12595986 |
| [27] |
doi: 10.1128/JVI.00955-10 URL |
| [28] |
pmid: 10811941 |
| [29] |
doi: 10.1186/s13059-022-02685-z pmid: 35597968 |
| [30] |
doi: 10.1093/oxfordjournals.jhered.a023078 URL |
| [31] |
pmid: 12904214 |
| [32] |
doi: 10.1093/emboj/21.3.461 URL |
| [33] |
pmid: 14985783 |
| [34] |
doi: 10.1093/nar/8.19.4321 pmid: 7433111 |
| [35] |
pmid: 17444906 |
| [36] |
doi: 10.1093/emboj/cdg443 pmid: 12970195 |
| [37] |
|
| [38] |
doi: 10.1007/s001220051337 URL |
| [39] |
doi: 10.1186/1471-2229-7-1 URL |
| [40] |
doi: 10.1016/j.tplants.2006.08.008 URL |
| [41] |
doi: 10.1146/annurev-biochem-080320-110356 pmid: 33556282 |
| [42] |
doi: 10.3389/fmicb.2019.02764 pmid: 31866963 |
| [43] |
pmid: 15680426 |
| [44] |
doi: 10.1186/1471-2148-10-1 |
| [45] |
doi: 10.1093/molbev/msac240 URL |
| [46] |
doi: S0168-1702(17)30698-6 pmid: 29792903 |
| [1] | ZHOU Jinhua, BAI Lei, ZHANG Rui, GUO Huachun. The Effect of Reducing Soil Temperature with Straw Mulching Cultivation on Potato Growth [J]. Acta Horticulturae Sinica, 2025, 52(2): 453-466. |
| [2] | HUANG Xun, LIU Xia, DENG Linmei, WANG Xingguo, XU Yajin, YANG Yanli. Identification and Study on Its Biocontrol and Growth Promoting Properties of Bacillus velezensis 1X1Y Against Potato Common Scab [J]. Acta Horticulturae Sinica, 2025, 52(1): 229-246. |
| [3] | ZHONG Yang, QIN Yazhi, LUO Shuai, JING Yuling, WANG Wanxing, LI Guangcun, HU Xinxi, QIN Yuzhi, CHENG Xu, XIONG Xingyao. Effects of Peat Soil and Mushroom Residue Application on the Potato Root-Associated Microbiome and Yield [J]. Acta Horticulturae Sinica, 2024, 51(9): 2143-2154. |
| [4] | GONG Xiaoya, LI Xian, ZHOU Xingang, WU Fengzhi. Effect of Rhizosphere Microorganisms Induced by Potato-Onion on Tomato Root-Knot Nematode Disease [J]. Acta Horticulturae Sinica, 2024, 51(8): 1913-1926. |
| [5] | FAN Guoquan, YU Jiang, GAO Yanling, LI Qingquan, ZHANG Shu, YU Zhenhua. Effects of Sprouting,Cutting and Coating of Potato Seed on the Occurrence of Rhizoctonia solani Disease [J]. Acta Horticulturae Sinica, 2024, 51(5): 1151-1161. |
| [6] | QIANG Ran, ZHANG Dai, YANG Zhe, CHEN Mingyue, ZHAO Jing, ZHU Jiehua. Effects of Bacillus amyloliquefaciens‘L19’on Fusarium oxysporum Inhibition and Plants Growth Promotion in Potato [J]. Acta Horticulturae Sinica, 2024, 51(4): 875-892. |
| [7] | LU Jianzeng, ZHOU Liyan, HUANG Xinyi, WU Fengzhi, GAO Danmei. Effects of Carbon Pool Strength on Plant Growth and Potassium Uptake of Tomato Intercropped with Potato-Onion [J]. Acta Horticulturae Sinica, 2024, 51(11): 2620-2632. |
| [8] | LI Shuang, LIN Yongxin, QIN Junhong, LI Guangcun, JIN Liping, LIU Jiangang, BIAN Chunsong. Research Progress in Potato Nitrogen Diagnosis [J]. Acta Horticulturae Sinica, 2024, 51(10): 2449-2468. |
| [9] | WANG Rongyan, LI Qing, XU Ling, WANG Qiujie, SONG Yu, MU Changran, TANG Wei, HAO Dahai. Genetic Analysis of Chloroplast Genome Characteristics of Potato‘Cooperation 88’Selfing Population [J]. Acta Horticulturae Sinica, 2023, 50(8): 1649-1663. |
| [10] | GONG Danmin, CAI Linzhi, DING Ren, MA Yanyan, WANG Wanxin, XIONG Xingyao, HU Xinxi. Effects of Different Potassium Fertilizers on Growth and Absorption and Accumulation of Mineral Elements and Cadmium in Potato [J]. Acta Horticulturae Sinica, 2023, 50(6): 1332-1342. |
| [11] | TAN Jie, LIU Jiangang, LIU Ju, JIAN Yinqiao, LI Guangcun, JIN Liping, XU Jianfei. Cultivated Substrates Evaluation for in-situ Observation of Potato Tubers Using Computed Tomography [J]. Acta Horticulturae Sinica, 2023, 50(5): 1085-1094. |
| [12] | YIN Jian, ZHU Xijian, WU Yaoyao, LI Canhui, ZHANG Jinzhe. Targets Prediction and Function Analysis of Potato miR319 Gene Family [J]. Acta Horticulturae Sinica, 2023, 50(3): 583-595. |
| [13] | XIONG Wenwen, LIU Huimin, CHEN Kaiyuan, DU Juan, SONG Botao, JING Shenglin. Optimization of Gene Editing System Based on Heat-treated CRISPR/Cas9 Potato Plants [J]. Acta Horticulturae Sinica, 2023, 50(12): 2591-2600. |
| [14] | YANG Jiangwei, DUAN Xiaoqin, ZHANG Feiyan, ZHANG Li, DENG Yurong, WANG Xiaofeng, ZHENG Zhiyong, ZHANG Ning, SI Huaijun. Root Architecture Alteration by Down-regulation of the StIAA22 Gene Using Artificial MicroRNA in Potato [J]. Acta Horticulturae Sinica, 2023, 50(10): 2091-2103. |
| [15] | WANG Kuan, QI Lipan, WU Guili, FENG Yan, WANG Lei, YIN Jiang, LUO Yating, WANG Yan, LIU Chang, GONG Xuechen, and WANG Haijun. A New Potato Cultivar‘Beifang 002’of Early Maturity and Good Yield [J]. Acta Horticulturae Sinica, 2022, 49(S2): 141-142. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd