
Acta Horticulturae Sinica ›› 2024, Vol. 51 ›› Issue (1): 203-212.doi: 10.16420/j.issn.0513-353x.2023-0170
• Plant Protection • Previous Articles Next Articles
GUAN Xiayu1,2, CHEN Xihong3, GUO Jing3, LÜ Haoyang1, LIANG Chenyuan1, GAO Fangluan1,*( )
)
Received:2023-05-11
Revised:2023-08-30
Online:2024-01-25
Published:2024-01-16
Contact:
(E-mail:GUAN Xiayu, CHEN Xihong, GUO Jing, LÜ Haoyang, LIANG Chenyuan, GAO Fangluan. Genetic Variability and Host Adaption of Cymbidium Mosaic Virus[J]. Acta Horticulturae Sinica, 2024, 51(1): 203-212.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2023-0170
| 寄主 Host | 采样省份 Location | 分离物 Isolate | GenBank 登录号 Accession No. | 寄主 Host | 采样省份 Location | 分离物 Isolate | GenBank 登录号 Accession No. | |
|---|---|---|---|---|---|---|---|---|
| 国兰 Cymbidium sinense | 福建Fujian | CHN-FJZZ12 | DQ067878 | 蝴蝶兰 Phalaenopsis aphrodite | 河南Henan | CHN-HNZZ02 | KU873001 | |
| CHN-FJZZ13 | DQ067879 | 蕙兰C. faberi | 西藏Tibet | C-LZ02 | KP137368 | |||
| 湖北Hubei | CHN-HBWH21 | DQ067880 | C-LZ06 | KP137369 | ||||
| CHN-HBWH22 | DQ067881 | C-LZ14 | KP137370 | |||||
| 浙江Zhejiang | CHN-ZJHZ52 | DQ067882 | C-LZ18 | KP137371 | ||||
| CHN-ZJHZ61 | DQ067883 | C-LZ21 | KP137372 | |||||
| CHN-ZJHZ62 | DQ067884 | 石斛兰 Dendrobium nobile | 中国台湾 Taiwan,China | CyMV-1 * | OQ615774 | |||
| CHN-ZJHZ63 | DQ067885 | CyMV-2 * | OQ615782 | |||||
| CHN-ZJHZ67 | DQ067886 | CyMV-3 * | OQ615783 | |||||
| CHN-ZJHZ69 | DQ067887 | CyMV-4 * | OQ615784 | |||||
| CHN-ZJHZ612 | DQ067888 | CyMV-5 * | OQ615785 | |||||
| 蝴蝶兰 Phalaenopsis aphrodite | 浙江Zhejiang | CyMV-hangzhou | AM398153 | CyMV-6 * | OQ615786 | |||
| CrMV-HZ | DQ915439 | CyMV-7 * | OQ615787 | |||||
| 福建Fujian | xm | GQ507023 | CyMV-8 * | OQ615788 | ||||
| 江苏Jiangsu | CrMV-NJ-1 | JQ860108 | CyMV-9 * | OQ615789 | ||||
| 广东Guangdong | FS2 | KF853247 | CyMV-10 * | OQ615775 | ||||
| FS3 | KF853248 | CyMV-11 * | OQ615776 | |||||
| FS4 | KF853249 | CyMV-12 * | OQ615777 | |||||
| FS5 | KF853250 | CyMV-13* | OQ615778 | |||||
| GZ1 | KF853251 | CyMV-14 * | OQ615779 | |||||
| CH | KF853252 | CyMV-15 * | OQ615780 | |||||
| GZ2 | KF853253 | CyMV-16 * | OQ615781 | |||||
| GZ3 | KF853254 | 广东Guangdong | ZHD1 | EU672821 | ||||
| FS6 | KF853255 | 建兰 C. ensifolium | 浙江Zhejiang | ZJXTHC1 | GU295167 | |||
| ZH1 | KF853256 | 四川Sichuan | SCJJC7 | GU295168 | ||||
| ZH2 | KF853257 | SCDXZC4 | GU295172 | |||||
| FS7 | KF853258 | 广东 Guangdong | GZKTLC6 | GU295170 | ||||
| FS8 | KF853259 | GDDHLC5 | GU295173 | |||||
| FS9 | KF853260 | 北京 Beijing | BJKTLC3 | GU295169 | ||||
| FS10 | KF853261 | 云南 Yunnan | KMMLC2 | GU295171 | ||||
| SZ1 | KF853262 | 墨兰 C. haematodes | 广东 Guangdong | gd2 | AY360409 | |||
| SZ2 | KF853263 | gd3 | AY360410 | |||||
| SZ3 | KF853264 | 中国台湾 Taiwan,China | CrMV-CS | AY429021 | ||||
| SZ4 | KF853265 | Taiwan,China | AY571289 | |||||
Table 1 CymMV isolates used in this study
| 寄主 Host | 采样省份 Location | 分离物 Isolate | GenBank 登录号 Accession No. | 寄主 Host | 采样省份 Location | 分离物 Isolate | GenBank 登录号 Accession No. | |
|---|---|---|---|---|---|---|---|---|
| 国兰 Cymbidium sinense | 福建Fujian | CHN-FJZZ12 | DQ067878 | 蝴蝶兰 Phalaenopsis aphrodite | 河南Henan | CHN-HNZZ02 | KU873001 | |
| CHN-FJZZ13 | DQ067879 | 蕙兰C. faberi | 西藏Tibet | C-LZ02 | KP137368 | |||
| 湖北Hubei | CHN-HBWH21 | DQ067880 | C-LZ06 | KP137369 | ||||
| CHN-HBWH22 | DQ067881 | C-LZ14 | KP137370 | |||||
| 浙江Zhejiang | CHN-ZJHZ52 | DQ067882 | C-LZ18 | KP137371 | ||||
| CHN-ZJHZ61 | DQ067883 | C-LZ21 | KP137372 | |||||
| CHN-ZJHZ62 | DQ067884 | 石斛兰 Dendrobium nobile | 中国台湾 Taiwan,China | CyMV-1 * | OQ615774 | |||
| CHN-ZJHZ63 | DQ067885 | CyMV-2 * | OQ615782 | |||||
| CHN-ZJHZ67 | DQ067886 | CyMV-3 * | OQ615783 | |||||
| CHN-ZJHZ69 | DQ067887 | CyMV-4 * | OQ615784 | |||||
| CHN-ZJHZ612 | DQ067888 | CyMV-5 * | OQ615785 | |||||
| 蝴蝶兰 Phalaenopsis aphrodite | 浙江Zhejiang | CyMV-hangzhou | AM398153 | CyMV-6 * | OQ615786 | |||
| CrMV-HZ | DQ915439 | CyMV-7 * | OQ615787 | |||||
| 福建Fujian | xm | GQ507023 | CyMV-8 * | OQ615788 | ||||
| 江苏Jiangsu | CrMV-NJ-1 | JQ860108 | CyMV-9 * | OQ615789 | ||||
| 广东Guangdong | FS2 | KF853247 | CyMV-10 * | OQ615775 | ||||
| FS3 | KF853248 | CyMV-11 * | OQ615776 | |||||
| FS4 | KF853249 | CyMV-12 * | OQ615777 | |||||
| FS5 | KF853250 | CyMV-13* | OQ615778 | |||||
| GZ1 | KF853251 | CyMV-14 * | OQ615779 | |||||
| CH | KF853252 | CyMV-15 * | OQ615780 | |||||
| GZ2 | KF853253 | CyMV-16 * | OQ615781 | |||||
| GZ3 | KF853254 | 广东Guangdong | ZHD1 | EU672821 | ||||
| FS6 | KF853255 | 建兰 C. ensifolium | 浙江Zhejiang | ZJXTHC1 | GU295167 | |||
| ZH1 | KF853256 | 四川Sichuan | SCJJC7 | GU295168 | ||||
| ZH2 | KF853257 | SCDXZC4 | GU295172 | |||||
| FS7 | KF853258 | 广东 Guangdong | GZKTLC6 | GU295170 | ||||
| FS8 | KF853259 | GDDHLC5 | GU295173 | |||||
| FS9 | KF853260 | 北京 Beijing | BJKTLC3 | GU295169 | ||||
| FS10 | KF853261 | 云南 Yunnan | KMMLC2 | GU295171 | ||||
| SZ1 | KF853262 | 墨兰 C. haematodes | 广东 Guangdong | gd2 | AY360409 | |||
| SZ2 | KF853263 | gd3 | AY360410 | |||||
| SZ3 | KF853264 | 中国台湾 Taiwan,China | CrMV-CS | AY429021 | ||||
| SZ4 | KF853265 | Taiwan,China | AY571289 | |||||
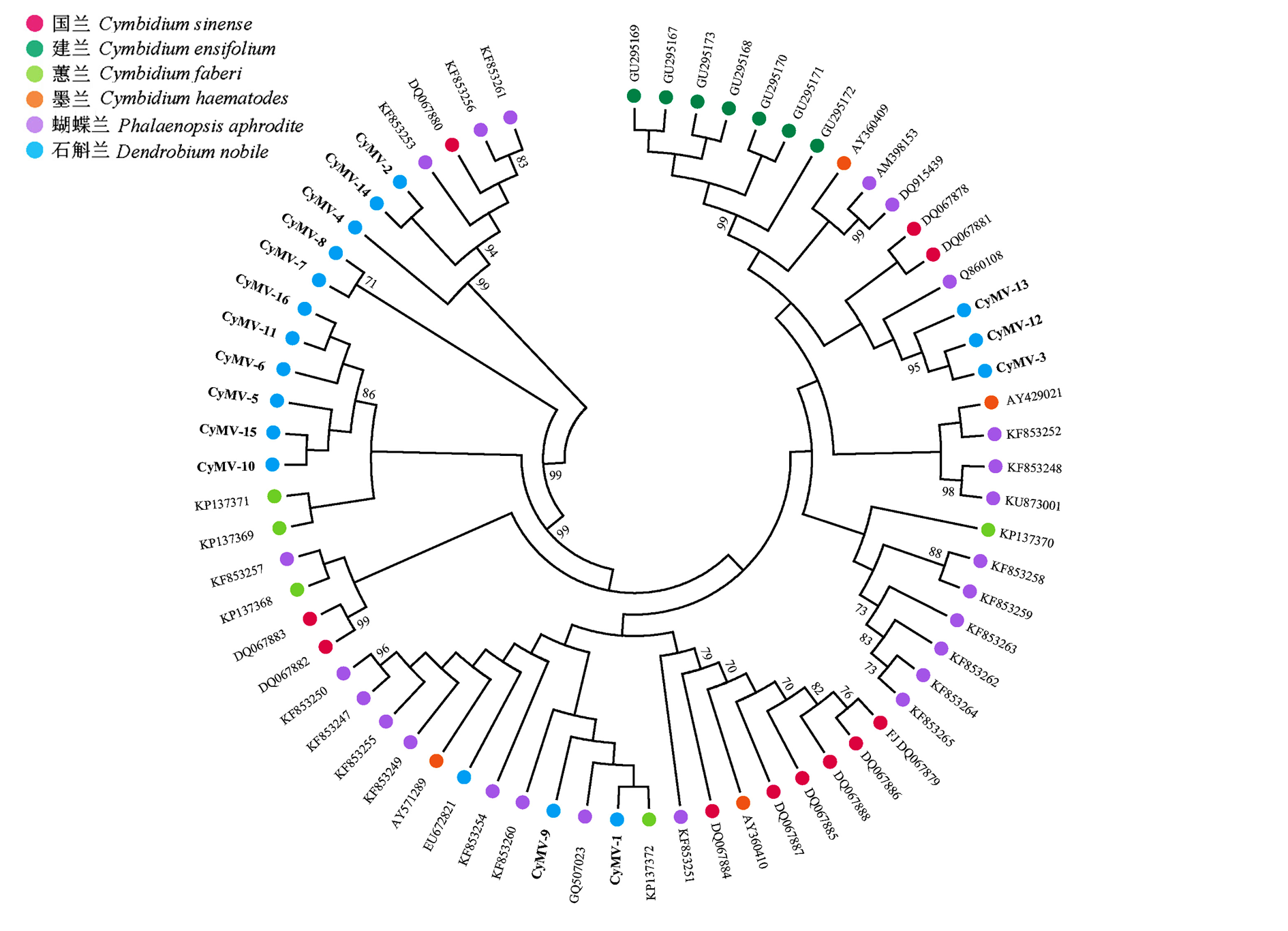
Fig. 3 Phylogenetic tree reconstructed from the CP gene sequence of CymMV Numbers above branches indicate the bootstrap values(only shown > 50%),and CymMV isolates from different host species are indicated by circles with differences in colours.
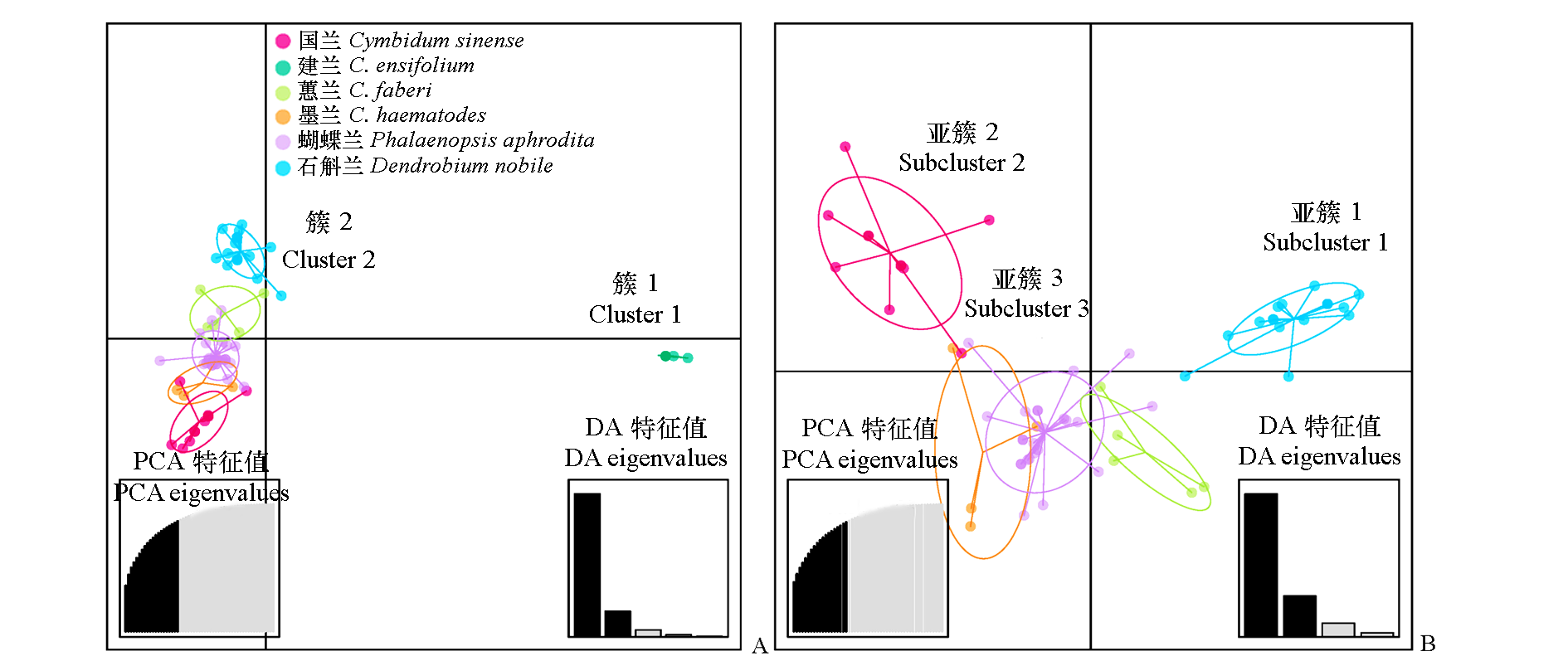
Fig. 4 Results from the discriminant analysis of principal components(DAPC) A:Scatterplots for 68 CymMV isolates; B:Scatterplots for CymMV isolates within Cluster 2.
| 簇 Cluster | 亚簇 Subcluster | 样本量大小 Sample size | 单倍型 Haplotypes | 单倍型多样性Hd Haplotype diversity | 核苷酸多样性Pi Nucleotide diversity |
|---|---|---|---|---|---|
| 1 | 7 | 3 | 0.524 ± 0.209 | 0.002 ± 0.009 | |
| 2 | 61 | 53 | 0.995 ± 0.004 | 0.040 ± 0.004 | |
| 2 | 1 | 17 | 14 | 0.971 ± 0.032 | 0.041 ± 0.008 |
| 2 | 11 | 9 | 0.961 ± 0.051 | 0.041 ± 0.012 | |
| 3 | 33 | 30 | 0.994 ± 0.009 | 0.036 ± 0.007 | |
| 总计Total | 68 | 56 | 0.992 ± 0.005 | 0.039 ± 0.004 |
Table 2 Summary statistics for genetic diversity of the CP gene of CymMV
| 簇 Cluster | 亚簇 Subcluster | 样本量大小 Sample size | 单倍型 Haplotypes | 单倍型多样性Hd Haplotype diversity | 核苷酸多样性Pi Nucleotide diversity |
|---|---|---|---|---|---|
| 1 | 7 | 3 | 0.524 ± 0.209 | 0.002 ± 0.009 | |
| 2 | 61 | 53 | 0.995 ± 0.004 | 0.040 ± 0.004 | |
| 2 | 1 | 17 | 14 | 0.971 ± 0.032 | 0.041 ± 0.008 |
| 2 | 11 | 9 | 0.961 ± 0.051 | 0.041 ± 0.012 | |
| 3 | 33 | 30 | 0.994 ± 0.009 | 0.036 ± 0.007 | |
| 总计Total | 68 | 56 | 0.992 ± 0.005 | 0.039 ± 0.004 |
| 变异源 Source of variation | 自由度 df | 平方和 Sum of squares | 变量组分 Variance components | 变异比例 Percentage of variation | 固定系数 Fixation index |
|---|---|---|---|---|---|
| 簇间Among clusters | 1 | 65.903 | 3.580 | 21.880 | 0.220 ns |
| 簇内亚簇间Among sub-clusters within clusters | 2 | 61.563 | 1.046 | 6.390 | 0.082* |
| 亚簇内Within sub-clusters | 64 | 750.931 | 11.733 | 71.720 | 0.282*** |
| 总计Total | 67 | 878.397 | 16.359 |
Table 3 Hierarchical analysis of molecular variance for the CP gene of CymMV
| 变异源 Source of variation | 自由度 df | 平方和 Sum of squares | 变量组分 Variance components | 变异比例 Percentage of variation | 固定系数 Fixation index |
|---|---|---|---|---|---|
| 簇间Among clusters | 1 | 65.903 | 3.580 | 21.880 | 0.220 ns |
| 簇内亚簇间Among sub-clusters within clusters | 2 | 61.563 | 1.046 | 6.390 | 0.082* |
| 亚簇内Within sub-clusters | 64 | 750.931 | 11.733 | 71.720 | 0.282*** |
| 总计Total | 67 | 878.397 | 16.359 |
| 簇 Cluster | 亚簇 Subcluster | Tajima's D | P值 P value | Fu's FS | P 值 P value |
|---|---|---|---|---|---|
| 1 | -1.434 | 0.067 | 0.263 | 0.475ns | |
| 2 | -1.335 | 0.043* | -19.765 | 0.000*** | |
| 2 | 1 | 0.483 | 0.729 | 0.029 | 0.517 ns |
| 2 | -1.446 | 0.072 | 1.339 | 0.696 ns | |
| 3 | -1.309 | 0.042* | -8.222 | 0.011* |
Table 4 Neutrality tests for different CymMV populations
| 簇 Cluster | 亚簇 Subcluster | Tajima's D | P值 P value | Fu's FS | P 值 P value |
|---|---|---|---|---|---|
| 1 | -1.434 | 0.067 | 0.263 | 0.475ns | |
| 2 | -1.335 | 0.043* | -19.765 | 0.000*** | |
| 2 | 1 | 0.483 | 0.729 | 0.029 | 0.517 ns |
| 2 | -1.446 | 0.072 | 1.339 | 0.696 ns | |
| 3 | -1.309 | 0.042* | -8.222 | 0.011* |
| 统计参数Statistic | 寄主 Host | 观测平均值(95% HPD) Observed mean(95% HPD) | 零假设平均值(95% HPD) Null mean(95% HPD) | P 值 P value |
|---|---|---|---|---|
| AI | 1.319(0.830 ~ 1.803) | 5.775(5.005 ~ 6.531) | < 0.001*** | |
| PS | 20.472(19.000 ~ 21.000) | 38.091(35.370 ~ 0.624) | < 0.001*** | |
| MC | 国兰Cymbidium sinense | 4.064(3.000 ~ 5.000) | 1.518(1.003 ~ 2.182) | 0.010** |
| 建兰C. ensifolium | 6.981(6.000 ~ 7.000) | 1.225(1.000 ~ 1.998) | 0.010** | |
| 蕙兰C. faberi | 2.049(2.000 ~ 3.000) | 1.103(1.000 ~ 1.989) | 0.030* | |
| 墨兰C. haematodes | 1.008(1.000 ~ 2.000) | 1.065(1.000 ~ 1.335) | 1.000ns | |
| 蝴蝶兰Phalaenopsis | 6.017(6.000 ~ 7.000) | 2.414(1.686 ~ 3.375) | 0.010** | |
| 石斛兰Dendrobium nobile | 5.975(5.000 ~ 6.000) | 1.887(1.079 ~ 2.982) | 0.010** |
Table 5 Bayesian Tip-association significance(BaTS)testing for the host effect on the genetic diversity of CymMV isolates
| 统计参数Statistic | 寄主 Host | 观测平均值(95% HPD) Observed mean(95% HPD) | 零假设平均值(95% HPD) Null mean(95% HPD) | P 值 P value |
|---|---|---|---|---|
| AI | 1.319(0.830 ~ 1.803) | 5.775(5.005 ~ 6.531) | < 0.001*** | |
| PS | 20.472(19.000 ~ 21.000) | 38.091(35.370 ~ 0.624) | < 0.001*** | |
| MC | 国兰Cymbidium sinense | 4.064(3.000 ~ 5.000) | 1.518(1.003 ~ 2.182) | 0.010** |
| 建兰C. ensifolium | 6.981(6.000 ~ 7.000) | 1.225(1.000 ~ 1.998) | 0.010** | |
| 蕙兰C. faberi | 2.049(2.000 ~ 3.000) | 1.103(1.000 ~ 1.989) | 0.030* | |
| 墨兰C. haematodes | 1.008(1.000 ~ 2.000) | 1.065(1.000 ~ 1.335) | 1.000ns | |
| 蝴蝶兰Phalaenopsis | 6.017(6.000 ~ 7.000) | 2.414(1.686 ~ 3.375) | 0.010** | |
| 石斛兰Dendrobium nobile | 5.975(5.000 ~ 6.000) | 1.887(1.079 ~ 2.982) | 0.010** |
| [1] |
|
|
常飞, 邹文超, 高芳銮, 沈建国, 詹家绥. 2015. 不同寄主来源的马铃薯Y病毒群体遗传结构的比较分析. 遗传, 37 (3): 292-301.
|
|
| [2] |
doi: 10.16420/j.issn.0513-353x.2020-1053 URL |
|
陈健鑫, 魏玉倩, 唐婕, 竺永金, 马焕成, 伍建榕. 2022. 云南大理栽培春兰中ORSV和CymMV的分子检测及序列分析. 园艺学报, 49 (3):655-662.
doi: 10.16420/j.issn.0513-353x.2020-1053 URL |
|
| [3] |
doi: 10.1099/vir.0.044347-0 pmid: 22837421 |
| [4] |
doi: 10.1111/j.1755-0998.2010.02847.x pmid: 21565059 |
| [5] |
|
| [6] |
doi: 10.1111/eva.2017.10.issue-4 URL |
| [7] |
doi: 10.3389/fmicb.2022.1051834 URL |
| [8] |
|
| [9] |
doi: 10.1016/j.ympev.2021.107336 URL |
| [10] |
doi: 10.1007/s11262-011-0592-x URL |
| [11] |
doi: 10.1186/1471-2156-11-94 URL |
| [12] |
doi: 10.1099/jgv.0.001436 pmid: 32525472 |
| [13] |
doi: 10.1093/molbev/msy096 URL |
| [14] |
doi: 10.16420/j.issn.0513-353x.2021-0886 URL |
|
李世帆, 邵宇纯, 祁豪男, 杨兰, 李小伟, 徐秉良, 陈雅寒. 2022. 甘肃省苹果坏死花叶病毒检测及遗传多样性分析. 园艺学报, 49 (11):2431-2438.
doi: 10.16420/j.issn.0513-353x.2021-0886 URL |
|
| [15] |
doi: 10.1093/bioinformatics/btp187 pmid: 19346325 |
| [16] |
doi: 10.1007/s00705-016-2773-3 pmid: 26923929 |
| [17] |
doi: 10.1093/ve/vev003 URL |
| [18] |
doi: 10.1007/s13205-019-1876-4 |
| [19] |
|
|
明艳林, 李梅, 郑国华. 2005. 建兰花叶病毒研究进展. 福建农业学报, 20 (1):30-33.
|
|
| [20] |
doi: 10.1007/s00705-006-0897-6 pmid: 17216134 |
| [21] |
doi: 10.1111/mpp.v21.2 URL |
| [22] |
doi: 10.1016/j.meegid.2007.08.001 URL |
| [23] |
doi: 10.1093/sysbio/sys029 pmid: 22357727 |
| [24] |
doi: 10.1093/molbev/msv222 pmid: 26503941 |
| [25] |
|
|
魏永路, 任锐, 高洁, 刘伟平, 谢琦, 朱根发, 杨凤玺. 2022. 建兰花叶病毒广东分离物的全基因组序列分析. 广东农业科学, 49 (2):109-117.
|
|
| [26] |
|
|
肖火根, 郑冠标, 张曙光, 高乔婉. 1996. 广东兰花病毒病的鉴定和检测研究. 华南农业大学学报, 17 (1):21-24.
|
|
| [27] |
|
|
谢联辉, 林奇英. 2011. 植物病毒学(第3版). 北京: 中国农业出版社.
|
|
| [28] |
doi: 10.1007/s11262-010-0468-5 URL |
| [29] |
|
| [30] |
doi: 10.3389/fmicb.2021.738646 URL |
| [1] | BI Qingrui, CUI Dongsheng, MA Xinyuan, XUE Yuran, ZHANG Shikui, FAN Guoquan, NIU Yingying. Genetic Diversity and Genetic Relationship of Local Pear Cultivars in Xinjiang [J]. Acta Horticulturae Sinica, 2025, 52(3): 561-574. |
| [2] | LUO Sifang, YAN Xiang, XIE Lifang, SUN Jingxian, GUO Zijing, ZHANG Zuming, CHEN Zhaoxing. Development and Application of Insertion-Deletion(InDel)Markers in Gannan wild Fortunella hindsii Based on Whole Genome Re-sequencing Data [J]. Acta Horticulturae Sinica, 2025, 52(2): 267-278. |
| [3] | ZHANG Lu, ZHANG Pingping, XIE Weijia, XU Feng, PENG Lüchun, SONG Jie, DU Guanghui, LI Shifeng. Genetic Diversity Analysis and SSR Fingerprint Construction of Evergreen Rhododendron Germplasms [J]. Acta Horticulturae Sinica, 2025, 52(2): 380-394. |
| [4] | XIAO Xi’ou, NIE Heng, LIN Wenqiu, WU Caiyu. Development of Whole-Genome SNP Markers of Eggplant [J]. Acta Horticulturae Sinica, 2025, 52(1): 88-100. |
| [5] | QIN Zilu, XU Zhengkang, DAI Xiaogang, CHEN Yingnan. Genetic Diversity Analysis and Core Collection Construction of Magnolia biondii Germplasm [J]. Acta Horticulturae Sinica, 2024, 51(8): 1823-1832. |
| [6] | YUAN Na, XU Qinyuan, XU Zhaolong, ZHOU Ling, LIU Xiaoqing, CHEN Xin, DU Jianchang. The Development and Verification of SNP Lquid Chips for Common Bean Based on Targeted Sequencing Technology [J]. Acta Horticulturae Sinica, 2024, 51(5): 1017-1032. |
| [7] | YOU Ping, YANG Jin, ZHOU Jun, HUANG Aijun, BAO Minli, YI Long. Genetic Diversity Analysis of Candidatus Liberibacter Asiaticus Based on Different Types of Prophage [J]. Acta Horticulturae Sinica, 2024, 51(4): 727-736. |
| [8] | XIONG Zhiwei, LI Zhilong, YIN Hui, GAO Yuxia. Pan-Genome Analysis of Candidatus Liberibacter asiaticus Causing Huanglongbing of Citrus [J]. Acta Horticulturae Sinica, 2024, 51(4): 737-747. |
| [9] | YOU Yuanyuan, WANG Shuai, FANG Yingying, QI Cheng, LI Shupei, ZHANG Ying, CHEN Yuhui, LIU Wei, LIU Fuzhong, SHU Jinshuai. Variation Characteristics of Insertion-Deletion(InDel)and Molecular Markers Development and Application in Eggplant Based on Whole Genome Re-sequencing Data [J]. Acta Horticulturae Sinica, 2024, 51(3): 520-532. |
| [10] | CHEN Fangce, HU Pingzheng, XIE Weiwei, WANG Zhengpeng, ZHONG Chuangnan, LI Haiyan, HE Yehua, PENG Ze, WAN Baoxiong, LIU Chaoyang. Development and Application of Prunus salicina InDel Markers Based on Genome Re-sequencing [J]. Acta Horticulturae Sinica, 2024, 51(11): 2510-2522. |
| [11] | WANG Rongbo, WANG Haiyun, ZHANG Qianrong, CAI Songling, LI Benjin, ZHANG Meixiang, LIU Peiqing. Genetic Diversity of Ralstonia solanacearum Strains Isolated from Fujian Province and Evaluation of Tomato Rootstocks for Resistance to Bacterial Wilt [J]. Acta Horticulturae Sinica, 2024, 51(11): 2540-2554. |
| [12] | LI Chunniu, SU Qun, LI Xianmin, HUANG Zhanwen, SUN Mingyan, LU Jiashi, WANG Hongyan, BU Zhaoyang. SSR Molecular Markers Development and Parentage Relationship Identification Based on Whole Genomic Sequences of Jasminum sambac [J]. Acta Horticulturae Sinica, 2024, 51(10): 2343-2357. |
| [13] | PEI Yun, YU Xiaqing, ZHAO Xiaokun, ZHANG Wanping, CHEN Jinfeng. Progress in the Study of the Association of Polyploidy with New Plant Phenotypes [J]. Acta Horticulturae Sinica, 2023, 50(9): 1854-1866. |
| [14] | WANG Rui, HONG Wenjuan, LUO Hua, ZHAO Lina, CHEN Ying, WANG Jun. Construction of SSR Fingerprints of Pomegranate Cultivars and Male Parent Identification of Hybrids [J]. Acta Horticulturae Sinica, 2023, 50(2): 265-278. |
| [15] | WANG Mengmeng, SUN Deling, CHEN Rui, YANG Yingxia, ZHANG Guan, LÜ Mingjie, WANG Qian, XIE Tianyu, NIU Guobao, SHAN Xiaozheng, TAN Jin, YAO Xingwei. Construction and Evaluation of Cauliflower Core Collection [J]. Acta Horticulturae Sinica, 2023, 50(2): 421-431. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd