
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (12): 3167-3179.doi: 10.16420/j.issn.0513-353x.2024-1034
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
CHEN Mengru, WU Qing’an, HE Sijia, CAI Xiangbin, MA Qiaoli, GU Qingqing, WEI Qingjiang*( )
)
Received:2025-02-17
Revised:2025-10-13
Online:2025-12-25
Published:2025-12-20
Contact:
WEI Qingjiang
CHEN Mengru, WU Qing’an, HE Sijia, CAI Xiangbin, MA Qiaoli, GU Qingqing, WEI Qingjiang. Genomic-Wide Identification and Expression Analysis of the DNA Methylation-Modifying Enzyme Genes in Poncirus trifoliata[J]. Acta Horticulturae Sinica, 2025, 52(12): 3167-3179.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-1034
| 基因 Gene | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer |
|---|---|---|
| PtrActin | CATCCCTCAGCACCTTCC | CCAACCTTAGCACTTCTCC |
| Pt5g021700 | AGGTTGT·GGTGGCTTATCTGATGG | TTCTCCTGCTGGTTGCTCATACTC |
| Pt7g014310 | TGATACCTTGGTGCCTGCCTAATAC | TGCCTTCCCAATCAAGCCTTCC |
| PtUn014750 | AGATTTAATACCCTGGTGCTTACC | AAAGTTTCCTTCCCAGTCCAA |
| PtUn014790 | TTATTTCCGCCCCAGGTTT | CTCAGGTGATGCTGCCCAT |
| PtUn014840 | CTTCTGCGAACATTTATACGATCC | TCCTGCTGGTTGCTCATACTCT |
| PtUn030910 | TGCCGCCTCACATCAAGA | CCATCAGATAAGCCACCACAA |
| Pt2g001260 | AAGTAAAGAAGCAGAAGCGAAGA | TCCCATCAACTGCCTCAAAC |
| Pt5g018630 | AGACGGTTCCTACAACAGCTACTTC | CGAGGCATAATCCAGTTGACATTCC |
| Pt5g023400 | AGCGAAGAAGAAGAAGAGGAGGAAG | TTCGGATCACCATAGCAGACAGC |
| Pt9g000560 | GTTGTCAGTCTTCAGCGGAATTGG | CCCACCACCTCTTGAGAATCCTTC |
| Pt1g006140 | GACATAGACCAGGAAGGCAAGGAG | ATGACAATCACCGCAAACACGAAG |
| Pt6g006730 | TGCGACTTCTAACACTCCACCATC | GCACCCAGCCGAGTCTTACAG |
| Pt8g006600 | GACTGTTGCCAGAACTGAGTATTGC | TGATCGCCTGTGCCTGTCTTG |
| Pt4g018910 | ACAACCGATGCTAGTGCGAGAC | TCTCTGCCTGCTCACCAATTACTTC |
Table 1 The sequences of qRT-PCR primers
| 基因 Gene | 正向引物序列(5′-3′) Forward primer | 反向引物序列(5′-3′) Reverse primer |
|---|---|---|
| PtrActin | CATCCCTCAGCACCTTCC | CCAACCTTAGCACTTCTCC |
| Pt5g021700 | AGGTTGT·GGTGGCTTATCTGATGG | TTCTCCTGCTGGTTGCTCATACTC |
| Pt7g014310 | TGATACCTTGGTGCCTGCCTAATAC | TGCCTTCCCAATCAAGCCTTCC |
| PtUn014750 | AGATTTAATACCCTGGTGCTTACC | AAAGTTTCCTTCCCAGTCCAA |
| PtUn014790 | TTATTTCCGCCCCAGGTTT | CTCAGGTGATGCTGCCCAT |
| PtUn014840 | CTTCTGCGAACATTTATACGATCC | TCCTGCTGGTTGCTCATACTCT |
| PtUn030910 | TGCCGCCTCACATCAAGA | CCATCAGATAAGCCACCACAA |
| Pt2g001260 | AAGTAAAGAAGCAGAAGCGAAGA | TCCCATCAACTGCCTCAAAC |
| Pt5g018630 | AGACGGTTCCTACAACAGCTACTTC | CGAGGCATAATCCAGTTGACATTCC |
| Pt5g023400 | AGCGAAGAAGAAGAAGAGGAGGAAG | TTCGGATCACCATAGCAGACAGC |
| Pt9g000560 | GTTGTCAGTCTTCAGCGGAATTGG | CCCACCACCTCTTGAGAATCCTTC |
| Pt1g006140 | GACATAGACCAGGAAGGCAAGGAG | ATGACAATCACCGCAAACACGAAG |
| Pt6g006730 | TGCGACTTCTAACACTCCACCATC | GCACCCAGCCGAGTCTTACAG |
| Pt8g006600 | GACTGTTGCCAGAACTGAGTATTGC | TGATCGCCTGTGCCTGTCTTG |
| Pt4g018910 | ACAACCGATGCTAGTGCGAGAC | TCTCTGCCTGCTCACCAATTACTTC |
| 基因名称 Gene name | 基因号 Gene ID | 编码序列长度/bp CDS Length | 氨基酸数 No. of amino acids | 分子量/D Molecular weight | 等电点 pI | 脂溶性指数 Aliphatic index | 不稳定系数 Instability index (II) | 亲水性 Gravy | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| PtrMET1-1 | Pt5g021700.1 | 5 247 | 1 748 | 196 887.65 | 6.18 | 78.07 | 40.18 | -0.439 | 细胞核 Nucleus |
| PtrMET1-2 | Pt7g014310.1 | 4 677 | 1 558 | 175 472.98 | 5.85 | 74.85 | 43.70 | -0.502 | 细胞核 Nucleus |
| PtrMET1-3 | PtUn014750.1 | 2 829 | 942 | 105 994.39 | 6.27 | 77.09 | 35.16 | -0.433 | 细胞质 Cytosol |
| PtrMET1-4 | PtUn014790.1 | 4 488 | 1 495 | 168 753.73 | 6.27 | 77.00 | 42.11 | -0.469 | 细胞核 Nucleus |
| PtrMET1-5 | PtUn014840.1 | 4 305 | 434 | 161 602.75 | 5.84 | 77.97 | 37.77 | -0.392 | 细胞核 Nucleus |
| PtrMET1-6 | PtUn030910.1 | 5 088 | 1 695 | 190 981.84 | 6.21 | 77.70 | 41.75 | -0.434 | 细胞核 Nucleus |
| PtrCMT1 | Pt2g001260.2 | 2 616 | 871 | 99 098.07 | 5.06 | 77.07 | 45.44 | -0.569 | 细胞核 Nucleus |
| PtrCMT2 | Pt5g018630.1 | 1 998 | 665 | 75 200.79 | 8.04 | 77.05 | 43.81 | -0.437 | 细胞核 Nucleus |
| PtrCMT3 | Pt5g023400.1 | 2 493 | 830 | 93 924.59 | 5.60 | 78.57 | 42.02 | -0.484 | 细胞核 Nucleus |
| PtrDRM | Pt9g000560.2 | 2 094 | 697 | 78 513.03 | 5.26 | 75.35 | 50.29 | -0.546 | 细胞核 Nucleus |
| PtrDNMT2 | Pt1g006140.1 | 3 543 | 1 180 | 132 606.24 | 5.80 | 70.49 | 39.30 | -0.580 | 细胞核 Nucleus |
| PtrROS1 | Pt6g006730.1 | 5 889 | 1 962 | 218 463.80 | 6.91 | 68.59 | 48.65 | -0.733 | 细胞核 Nucleus |
| PtrDML3 | Pt8g006600.1 | 5 250 | 1 749 | 196 115.50 | 8.25 | 73.01 | 54.29 | -0.689 | 细胞核 Nucleus |
| PtrDME | Pt4g018910.1 | 5 874 | 1 957 | 219 765.43 | 6.49 | 66.69 | 55.85 | -0.763 | 细胞核 Nucleus |
Table 2 Physicochemical properties of DNA methylation-modifying enzyme genes in Poncirus trifoliata
| 基因名称 Gene name | 基因号 Gene ID | 编码序列长度/bp CDS Length | 氨基酸数 No. of amino acids | 分子量/D Molecular weight | 等电点 pI | 脂溶性指数 Aliphatic index | 不稳定系数 Instability index (II) | 亲水性 Gravy | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| PtrMET1-1 | Pt5g021700.1 | 5 247 | 1 748 | 196 887.65 | 6.18 | 78.07 | 40.18 | -0.439 | 细胞核 Nucleus |
| PtrMET1-2 | Pt7g014310.1 | 4 677 | 1 558 | 175 472.98 | 5.85 | 74.85 | 43.70 | -0.502 | 细胞核 Nucleus |
| PtrMET1-3 | PtUn014750.1 | 2 829 | 942 | 105 994.39 | 6.27 | 77.09 | 35.16 | -0.433 | 细胞质 Cytosol |
| PtrMET1-4 | PtUn014790.1 | 4 488 | 1 495 | 168 753.73 | 6.27 | 77.00 | 42.11 | -0.469 | 细胞核 Nucleus |
| PtrMET1-5 | PtUn014840.1 | 4 305 | 434 | 161 602.75 | 5.84 | 77.97 | 37.77 | -0.392 | 细胞核 Nucleus |
| PtrMET1-6 | PtUn030910.1 | 5 088 | 1 695 | 190 981.84 | 6.21 | 77.70 | 41.75 | -0.434 | 细胞核 Nucleus |
| PtrCMT1 | Pt2g001260.2 | 2 616 | 871 | 99 098.07 | 5.06 | 77.07 | 45.44 | -0.569 | 细胞核 Nucleus |
| PtrCMT2 | Pt5g018630.1 | 1 998 | 665 | 75 200.79 | 8.04 | 77.05 | 43.81 | -0.437 | 细胞核 Nucleus |
| PtrCMT3 | Pt5g023400.1 | 2 493 | 830 | 93 924.59 | 5.60 | 78.57 | 42.02 | -0.484 | 细胞核 Nucleus |
| PtrDRM | Pt9g000560.2 | 2 094 | 697 | 78 513.03 | 5.26 | 75.35 | 50.29 | -0.546 | 细胞核 Nucleus |
| PtrDNMT2 | Pt1g006140.1 | 3 543 | 1 180 | 132 606.24 | 5.80 | 70.49 | 39.30 | -0.580 | 细胞核 Nucleus |
| PtrROS1 | Pt6g006730.1 | 5 889 | 1 962 | 218 463.80 | 6.91 | 68.59 | 48.65 | -0.733 | 细胞核 Nucleus |
| PtrDML3 | Pt8g006600.1 | 5 250 | 1 749 | 196 115.50 | 8.25 | 73.01 | 54.29 | -0.689 | 细胞核 Nucleus |
| PtrDME | Pt4g018910.1 | 5 874 | 1 957 | 219 765.43 | 6.49 | 66.69 | 55.85 | -0.763 | 细胞核 Nucleus |

Fig. 1 Phylogenetic analysis,chromosome localization,and syntenic analysis of DNA methylation-modifying enzyme genes A:Phylogenetic analysis of DNA methylation-modifying enzyme genes from Poncirus trifoliata(Ptr),Arabidopsis thaliana(At),and Oryza sativa(Os);B:Chromosome localization and syntenic analysis of DNA methylation-modifying enzyme genes in Poncirus trifoliata;C:Syntenic analysis of DNA methylation-modifying enzyme genes from Poncirus trifoliata,Arabidopsis thaliana,and Oryza sativa

Fig. 3 The type(A)and number(B)of cis-acting elements in the promoter region,and protein interaction network(C)of DNA methylation-modifying enzyme genes in Poncirus trifoliata
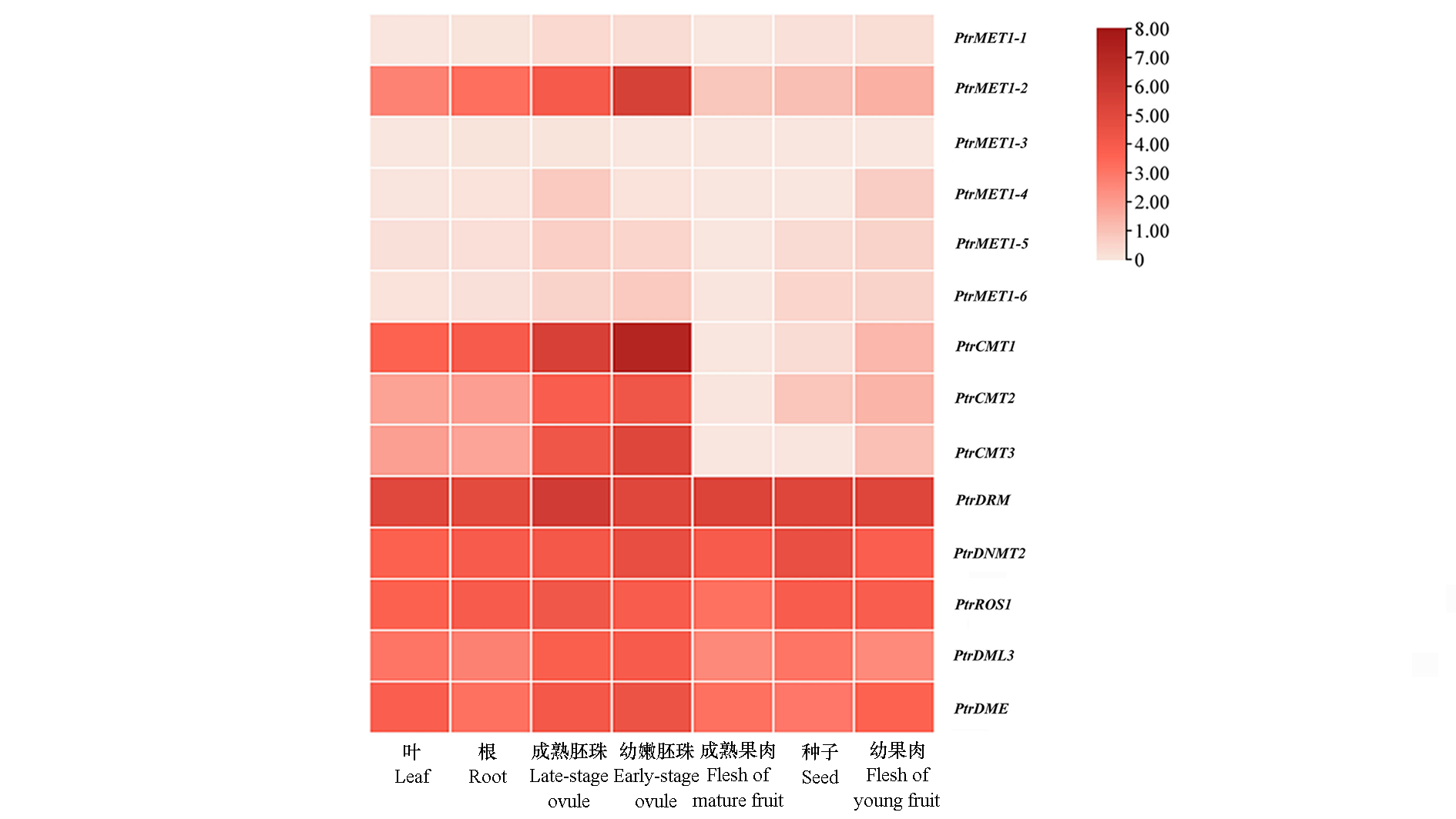
Fig. 4 Expression heatmap of DNA methylation-modifying enzyme genes in different tissues of Poncirus trifoliata The data in the heatmap shows the log2FPKM value
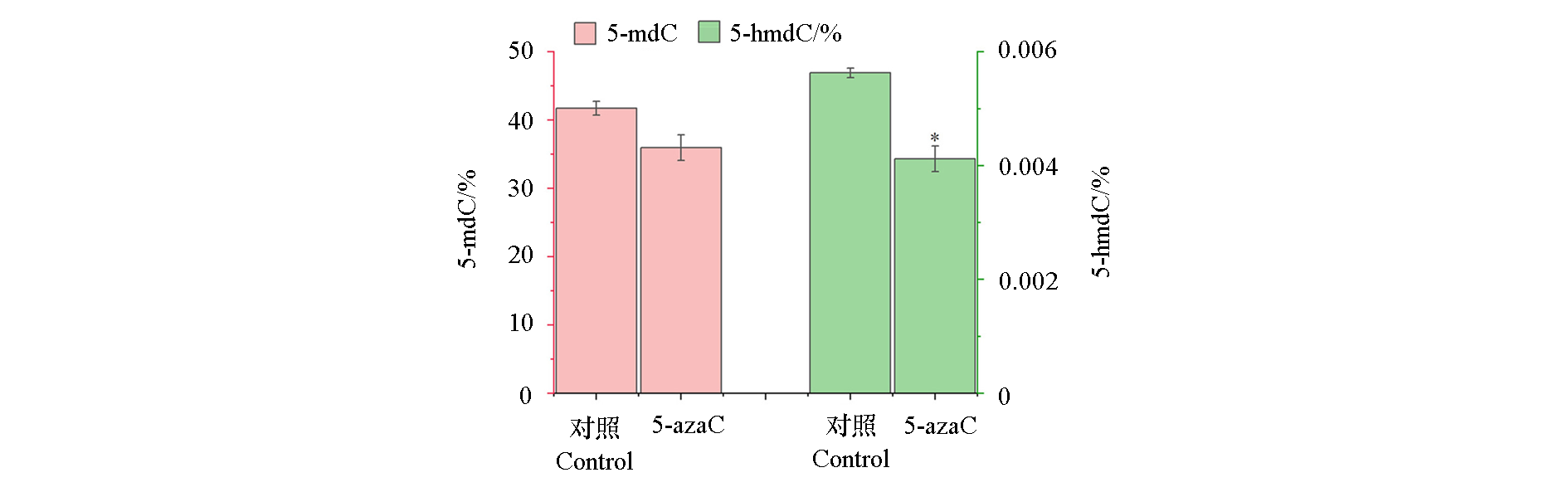
Fig. 5 DNA methylation level in leaf of Poncirus trifoliata after 5-azaC treatment Differences between the means were evaluated using Student’s t-test,and the * indicates significant difference between the control and 5-azaC treatment(P < 0.05)
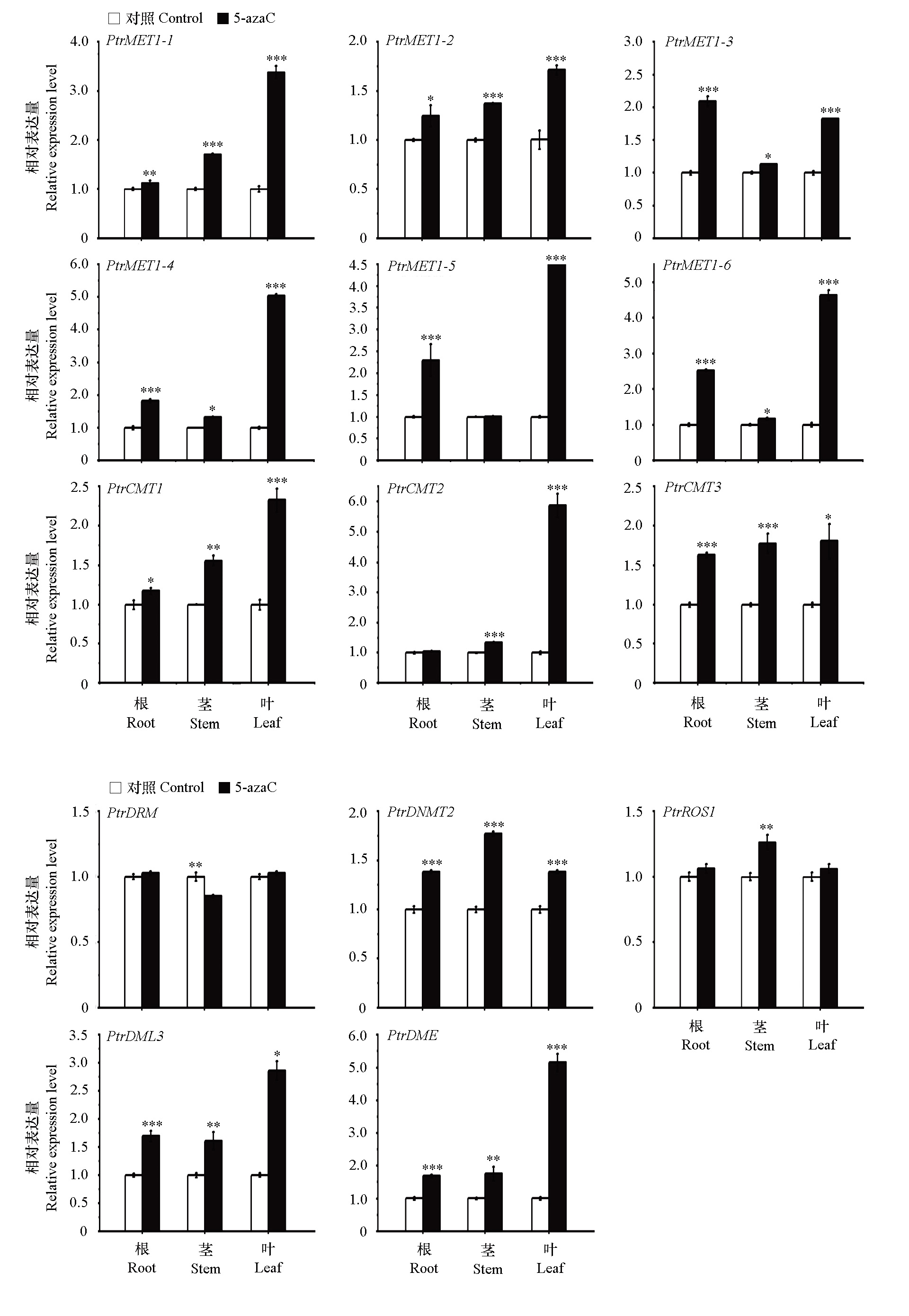
Fig. 6 Expression of DNA methylation-modifying enzyme genes in root,stem,and leaf of Poncirus trifoliata after 5-azaC treatment Differences between the means were evaluated using Student’s t-test,and the *,**,and *** indicate significant difference between the CK and 5-azaC treatment at P < 0.05,P < 0.01,and P < 0.001,respectively
| [35] |
|
| [36] |
|
|
徐记迪. 2015. 柑橘全基因组DNA甲基化分析及调控作用研究[博士论文]. 武汉: 华中农业大学.
|
|
| [37] |
|
| [38] |
|
|
杨惠栋, 彭震宇, 彭良志, 袁梦, 淳长品, 袁高鹏. 2023. 不同砧木对纽荷尔脐橙嫁接苗生长和生理指标的影响. 江西农业大学学报, 45(3):575-583.
|
|
| [39] |
|
| [40] |
|
|
杨志宁, 陆许可, 范亚朋, 孙玉平, 喻欣, 王亮, 高永健, 古丽加依那什·哈不力, 古丽沙西·拿斯依, 吾木尔·阿不都, 黄慧, 张梦豪, 王立东, 陈筱, 肖磊, 张鑫蕊, 王帅, 陈修贵, 王俊娟, 郭丽雪, 高文伟, 叶武威. 2025. 棉花GhDMT7基因对5-氮杂胞嘧啶核苷的响应机制. 中国农业科技导报, 27(8):28-35.
|
|
| [41] |
|
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
|
陈婷欣, 符敏, 李娜, 杨蕾蕾, 李凌飞, 钟春梅. 2024. 铁甲秋海棠DNA甲基转移酶全基因组鉴定及表达分析. 植物学报, 59(5):726-737.
|
|
| [10] |
|
| [11] |
|
|
邓秀新. 2022. 中国柑橘育种60年回顾与展望. 园艺学报, 49(10):2063-2074.
|
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [42] |
|
|
张盼, 余永旭, 曹领改, 朱广兵, 吴伟, 郭玉双, 尹国英, 贾蒙骜. 2023. m6A甲基化修饰响应植物生物胁迫和非生物胁迫的研究进展. 园艺学报, 50(9):1841-1853.
|
|
| [43] |
|
| [44] |
|
| [45] |
|
|
朱世平, 王福生, 陈娇, 余歆, 余洪, 罗国涛, 胡洲, 冯锦英, 赵晓春, 洪棋斌. 2020. 柑橘不同类型砧木的种子和苗期性状. 中国农业科学, 53(3):585-599.
|
|
| [46] |
|
|
庄泽岳. 2023. 辣椒DRM基因家族的分析及CaDRM1在花器官发育中的功能研究[硕士论文]. 广州: 仲恺农业工程学院.
|
|
| [29] |
|
|
马青龄, 梁贝贝, 杨莉, 胡威, 刘勇, 刘德春. 2022. 枳WOX基因家族全基因组鉴定及表达分析. 果树学报, 39(5):712-729.
|
|
| [30] |
|
| [31] |
|
| [32] |
|
|
祁春节, 顾雨檬, 曾彦. 2021. 我国柑橘产业经济研究进展. 华中农业大学学报, 40(1):58-69.
|
|
| [33] |
|
| [34] |
|
| [1] | TANG Jiayi, MIAO Chouyu, YIN Haojie, WU Mingyue, CAO Xiangmin, ZHANG Huimin, JIN Yan, LU Xiaopeng, ZHU Yichi, LI Dazhi, SHENG Ling. The Effect of Exogenous Oxalic Acid Treatment on the Fruit Quality and Anthocyanin Metabolism of Postharvest Blood Orange Fruit [J]. Acta Horticulturae Sinica, 2025, 52(9): 2425-2438. |
| [2] | DOU Xueting, ZHU Xi, ZHANG Ning, and SI Huaijun. Functional Analysis of StHY5 Associated with Low-Temperature Stress in Potato [J]. Acta Horticulturae Sinica, 2025, 52(7): 1745-1757. |
| [3] | ZHOU Tian, LÜ Shuaili, HAN Han, LI Yujia, LIU Haiqiang, and LIU Yongzhong. Effect of Exogenous Strigolactones on the Secondary Growth of Poncirus trifoliata Seedlings Stem [J]. Acta Horticulturae Sinica, 2025, 52(7): 1843-1854. |
| [4] | WANG Yi, LIANG Zhenchang. Analysis of Genetic Mechanism of Grape Distant Hybridization Based on Population Transcriptome [J]. Acta Horticulturae Sinica, 2025, 52(4): 835-845. |
| [5] | TANG Lingli, HE Yuhua, WANG Qingtao, AN Lulu, XU Yongyang, ZHANG Jian, KONG Weihu, HU Keyun, ZHAO Guangwei. Hormonal and Transcriptome Analysis of Ripe Fruits of Non-Climacteric and Climacteric Melon [J]. Acta Horticulturae Sinica, 2025, 52(4): 883-896. |
| [6] | SUN Tingzhen, SHU Qin, MA Wei, SHI Yuzi, ZHANG Meng, XIANG Chenggang, BO Kailiang, DUAN Ying, WANG Changlin. Identification and Phylogenetic Analysis of Gibberellin Oxidase Gene Family and Its Expression Pattern in Main Vine Growth in Cucurbita maxima [J]. Acta Horticulturae Sinica, 2025, 52(3): 603-622. |
| [7] | WANG Jinxuan, NI Ying, MENG Xin, LENG Zhuo, MA Bo, LENG Pingsheng, WU Jing, HU Zenghui. Functional Analysis of SoCHR35 in the Biosynthesis of Ocimene in Syringa oblata [J]. Acta Horticulturae Sinica, 2025, 52(10): 2655-2665. |
| [8] | LI Hui, KAN Jialiang, LI Xiaogang, WANG Zhonghua, LIN Jing, YANG Qingsong. Effect of Gibberellin on Wizened Flower Buds Formation of Pyrus pyrifolia Cultivar‘Sucui 1’During the Growing Season [J]. Acta Horticulturae Sinica, 2025, 52(10): 2713-2726. |
| [9] | ZHOU Xiaolin, LIN Junxuan, LI Caixia, SUN Meihua, BAI Longqiang, HOU Leiping, MIAO Yanxiu. Effects of Exogenous Glutathione on Photosynthetic Characteristics of Leaf Lettuce Under High Temperature Stress [J]. Acta Horticulturae Sinica, 2025, 52(10): 2761-2773. |
| [10] | HE Dandan, HE Hongtai, WANG Wenting, ZHOU Wenmei, LIU Yanmin, LIU Sushuang. Identification of Melon GolS Genes Family and the Expression Analysis in Response to Low Temperature Stress [J]. Acta Horticulturae Sinica, 2025, 52(1): 136-148. |
| [11] | SHI Xingxiu, FENG Beibei, YAN Peng, GENG Wenjuan, Jumazira Sharshanmukhan. Physiological Changes Associated with Early Watercore in‘Orin’Apples [J]. Acta Horticulturae Sinica, 2025, 52(1): 171-184. |
| [12] | REN Siyuan, CHEN Sen, LONG Zhijian, WANG Boya, TANG Dengguo, WANG Zhengqian, YANG Bin, HU Shanglian, CAO Ying. Nitrogen Allocation Characteristics and Expression of Related Genes During Corm-Forming Stage of Amorphophallus konjac [J]. Acta Horticulturae Sinica, 2024, 51(9): 2019-2030. |
| [13] | ZHOU Ping, YAN Shaobin, GUO Rui, JIN Guang. Identification and Expressional Analysis of MGT Gene Family in Prunus persica [J]. Acta Horticulturae Sinica, 2024, 51(3): 463-478. |
| [14] | YAN Chaofan, SUN Xuemei, ZHONG Qiwen, SHAO Dengkui, DENG Changrong, WEN Junqin. Identification and Bioinformatics Analysis of 20S Proteasome Gene Family in Tomato [J]. Acta Horticulturae Sinica, 2024, 51(2): 266-280. |
| [15] | CUI Yiqiong, LI Ju, LIU Xiaoqi, WANG Junwen, TANG Zhongqi, WU Yue, XIAO Xuemei, YU Jihua. Effects of Water Deficiency on Sucrose and Starch Metabolism of Substrate Culture Tomato in Greenhouse [J]. Acta Horticulturae Sinica, 2024, 51(11): 2607-2619. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd