
园艺学报 ›› 2025, Vol. 52 ›› Issue (10): 2597-2612.doi: 10.16420/j.issn.0513-353x.2024-0850
杨敏*, 贺涵*, 周陈平, 邝瑞彬, 刘传和, 吴夏明, 徐泽, 魏岳荣**( )
)
收稿日期:2025-03-02
修回日期:2025-07-28
出版日期:2025-10-25
发布日期:2025-10-28
通讯作者:
作者简介:*共同第一作者
基金资助:
YANG Min, HE Han, ZHOU Chenping, KUANG Ruibin, LIU Chuanhe, WU Xiaming, XU Ze, WEI Yuerong**( )
)
Received:2025-03-02
Revised:2025-07-28
Published:2025-10-25
Online:2025-10-28
摘要:
利用5份番木瓜种质的重测序数据挖掘InDel(插入/缺失)位点,并基于位点设计高效且具有应用价值的分子标记。经严格筛选后,成功鉴定3 635个易于凝胶电泳检测、长度大于30 bp且呈现高度多态性的InDel位点。挑选360个在基因组上均匀分布的位点设计引物,用6份番木瓜材料初筛后发现355对引物能成功扩增出目标产物,其中271对表现出显著的多态性,占候选引物总数的75.3%。从中选取72对高多态性标记对47份番木瓜种质进行基因分型和遗传多样性分析,共检测到145个等位位点;标记的多态性信息含量(PIC)分布于0.0408 ~ 0.4523,平均值为0.3041;基因多样性指数的变异范围为0.1030 ~ 0.8735,平均值为0.5660。通过聚类分析,将47份番木瓜划分为4个类群。此外,基于20对高多态性核心InDel标记的扩增结果,结合花色、单果质量、果形和果肉颜色4种关键农艺性状,构建番木瓜种质的分子身份证。同时,选取能反映亲本间差异的两个InDel标记,鉴定24份番木瓜F1(GT1-5 ב黄金钟’)杂交后代的真实性,成功确认11个真实杂交种。
杨敏, 贺涵, 周陈平, 邝瑞彬, 刘传和, 吴夏明, 徐泽, 魏岳荣. 基于重测序的番木瓜基因组InDel标记开发与应用[J]. 园艺学报, 2025, 52(10): 2597-2612.
YANG Min, HE Han, ZHOU Chenping, KUANG Ruibin, LIU Chuanhe, WU Xiaming, XU Ze, WEI Yuerong. Development and Application of Insertion-Deletion(InDel)Markers in Papaya(Carica papaya)Based on Whole Genome Re-Sequencing Data[J]. Acta Horticulturae Sinica, 2025, 52(10): 2597-2612.
| 染色体编号 Chrosome labels | 标记引物数 Number of InDel primers | 有效引物数 Number of effective primers | 有效扩增率/% Effective amplification ratios | 多态引物数 Number of polymorphic primers | 引物多态率/% Polymorphic primer ratios |
|---|---|---|---|---|---|
| Chr1 | 40 | 36 | 90.0 | 31 | 77.5 |
| Chr2 | 40 | 40 | 100.0 | 34 | 85.0 |
| Chr3 | 40 | 39 | 97.5 | 31 | 77.5 |
| Chr4 | 40 | 40 | 100.0 | 32 | 80.0 |
| Chr5 | 40 | 40 | 100.0 | 28 | 70.0 |
| Chr6 | 40 | 40 | 100.0 | 27 | 67.5 |
| Chr7 | 40 | 40 | 100.0 | 28 | 70.0 |
| Chr8 | 40 | 40 | 100.0 | 29 | 72.5 |
| Chr9 | 40 | 40 | 100.0 | 31 | 77.5 |
| 合计Total | 360 | 355 | 98.6 | 271 | 75.3 |
表1 360对InDel标记引物的扩增结果统计
Table 1 Statistics of amplification results of 360 InDel markers
| 染色体编号 Chrosome labels | 标记引物数 Number of InDel primers | 有效引物数 Number of effective primers | 有效扩增率/% Effective amplification ratios | 多态引物数 Number of polymorphic primers | 引物多态率/% Polymorphic primer ratios |
|---|---|---|---|---|---|
| Chr1 | 40 | 36 | 90.0 | 31 | 77.5 |
| Chr2 | 40 | 40 | 100.0 | 34 | 85.0 |
| Chr3 | 40 | 39 | 97.5 | 31 | 77.5 |
| Chr4 | 40 | 40 | 100.0 | 32 | 80.0 |
| Chr5 | 40 | 40 | 100.0 | 28 | 70.0 |
| Chr6 | 40 | 40 | 100.0 | 27 | 67.5 |
| Chr7 | 40 | 40 | 100.0 | 28 | 70.0 |
| Chr8 | 40 | 40 | 100.0 | 29 | 72.5 |
| Chr9 | 40 | 40 | 100.0 | 31 | 77.5 |
| 合计Total | 360 | 355 | 98.6 | 271 | 75.3 |
| 标记号 InDel labels | 标记类型 Type | 差异长 度/bp Length | 上游扩增引物(5′-3′) Forward primer | 下游扩增引物(5′-3′) Reverse primer | 标记引物扩增位置 Amplification location |
|---|---|---|---|---|---|
| Chr1-09 | 插入 Insertion | 35 | ACCTTGACATACATGCATGCA | TGCCTTGAGTCACACGGA | Chr1:2749817-2750039 |
| Chr1-10 | 插入 Insertion | 36 | GAACCTCCTCCCCCGCTA | TGAACCACCTGCTCGCAT | Chr1:2857128-2857366 |
| Chr1-15 | 插入 Insertion | 49 | TGTTCCATGTGTTATGGACGT | ACAAGCTTGGTGCATATGCT | Chr1:3213152-3213384 |
| Chr1-26 | 缺失 Deletion | -46 | CGCCCTCTCCCGTAAAGC | ACCACAGGTACTTGGCCG | Chr1:7259239-7259506 |
| Chr1-29 | 缺失 Deletion | -30 | CGTGAGGTGCTCGCTGAT | GGGACATCTCGTGCACGT | Chr1:7846667-7846868 |
| Chr1-30 | 插入 Insertion | 42 | GAACGCAAAAGACTGACTCCT | AGGTTCTGAAGACATGCACTT | Chr1:8015769-8016044 |
| Chr1-39 | 插入 Insertion | 39 | GTCCTCAGGACGACCCTAGA | ATCTGGTGGCAGTGGCAG | Chr1:10631445-10631724 |
| Chr2-01 | 插入 Insertion | 37 | GCTCCAACGAGAGAGATGACT | CGGTGCTGGTGACGAAGA | Chr2:307044-307320 |
| Chr2-02 | 插入 Insertion | 31 | CAGGATTTTGCTAGGGTCAGT | AGACTTGCCTGCTGGAGC | Chr2:612393-612613 |
| Chr2-13 | 缺失 Deletion | -34 | ACCAACCGTCGATCCTCA | TGGCAAAACGTCGTGATGT | Chr2:5215597-5215796 |
| Chr2-15 | 插入 Insertion | 44 | TGCTGGGAATGCTTTGTGC | TGGTCCATGATGCCAGCTG | Chr2:5922591-5922860 |
| Chr2-18 | 插入 Insertion | 44 | GGTTAGACTGTCAAGTGGGCT | CGCCCTGAAAACATGGTGC | Chr2:10719434-10719712 |
| Chr2-20 | 缺失 Deletion | -31 | ACAACCTGAGAGCTCTTCCC | AGCTGCTTCTACCTGCTTGA | Chr2:12083278-12083527 |
| Chr2-29 | 插入 Insertion | 48 | TGGAATCAAGATTTGCACCGA | TTCCAGTTTCCGAACAACCA | Chr2:23526410-23526648 |
| Chr2-31 | 缺失 Deletion | -33 | GGCAGCACTTCATTGCCG | GGGTCTTACACTGGTTGCCA | Chr2:28941802-28942025 |
| Chr2-38 | 插入 Insertion | 38 | ACGAGTCTTCAAGCTTGCCA | TGCCAGTCTGTAAATTGCGG | Chr2:34031300-34031578 |
| Chr3-03 | 插入 Insertion | 48 | GCCAACACAAGAACGGAAACT | TGCAGTTCAAGTGGCGCT | Chr3:627145-627348 |
| Chr3-17 | 缺失 Deletion | -44 | CTCCTGCACAGTCGCCAT | GCTTGCTTTGCTCTCCGC | Chr3:6901713-6901929 |
| Chr3-21 | 缺失 Deletion | -46 | CCCTTCCCCCTCCCAAGA | TTCTGCCGTCGGTTGAGC | Chr3:10826323-10826544 |
| Chr3-22 | 插入 Insertion | 39 | TCCCGGACATCTGACCCC | AGCCTTTGATTCCCACTGCA | Chr3:10886342-10886562 |
| Chr3-23 | 插入 Insertion | 31 | GGAGCACAAGCCATGGGT | AGCCACCAGTCAGACCTCT | Chr3:11351904-11352128 |
| Chr3-30 | 插入 Insertion | 34 | ACGGTGACAGCATGAGGC | CACATGCAAGATGCCGGC | Chr3:16193105-16193338 |
| Chr3-31 | 插入 Insertion | 42 | AGCCAACCCACCACAAGT | CTCAGCCTATTTGTTGGTCCA | Chr3:16707449-16707694 |
| Chr3-33 | 插入 Insertion | 33 | TGCTGGAAAACACCCGCA | GCCTCCCCAAGTCAGCTG | Chr3:21024394-21024593 |
| Chr4-03 | 缺失 Deletion | -33 | CCAGCATCAGAACTCGAGGT | TTGGTTGCGGGGAATGCA | Chr4:1324489-1324712 |
| Chr4-08 | 插入 Insertion | 42 | CGAGCCTGTGTACCTGGT | TCACTTTCGTCAGTTGCCTGA | Chr4:4076944-4077153 |
| Chr4-14 | 插入 Insertion | 41 | GCGGTGAGCCTGAGATGA | AGACTCGTGGAGAACAAAGCA | Chr4:9192243-9192474 |
| Chr4-24 | 缺失 Deletion | -40 | ACGGGGCGTACATGTGTG | ACGAGGCCACCCCTAAGA | Chr4:21621919-21622128 |
| Chr4-25 | 插入 Insertion | 45 | TGGTTTCGCAAGTTCGCA | GTGCTAGTTGTTTTGCATGCT | Chr4:22997264-22997514 |
| Chr4-26 | 缺失 Deletion | -46 | AGAGGGGTTAGGCAGGGG | TCCCCACATTGATGTTGACCA | Chr4:23149465-23149734 |
| Chr4-34 | 缺失 Deletion | -32 | TCACGAGTTGAGGGAGGTCT | ACTTGTTGAGGGTTGAAAGCC | Chr4:28955964-28956194 |
| Chr4-40 | 插入 Insertion | 46 | GACGTGGTGCTTATCCCGT | GGCAGTCCAGTTCTATAGGGG | Chr4:34041449-34041703 |
| Chr5-02 | 缺失 Deletion | -49 | TGCGCACAACATTCAGGTC | GGCTATGTTGTGCCTGGGA | Chr5:59655-59828 |
| Chr5-07 | 插入 Insertion | 38 | TGAGTGGAAAGAAGATGCGGA | TCATCCCTCCATCCACAAACT | Chr5:272642-272800 |
| Chr5-08 | 缺失 Deletion | -34 | GACAAAGCAAAACTGCATCGT | TGATTTGGCAGCCCCTGG | Chr5:699745-699920 |
| Chr5-11 | 缺失 Deletion | -35 | GCGGCAATCAGAGGAGCA | GCACCACACAGACAAAAATCA | Chr5:1024290-1024446 |
| Chr5-15 | 缺失 Deletion | -33 | TGGGAGGTAGCTGACGGA | CAGCATGCCTAACCGTGC | Chr5:1509285-1509412 |
| Chr5-18 | 缺失 Deletion | -33 | GACTCCGATGATGTCCTCTGT | CGTGAAGGAAATCGGTGTGC | Chr5:1615976-1616100 |
| Chr5-24 | 插入 Insertion | 31 | GGGCTTGAGTTTGGGCCT | CCCAGTCCCATACCCCGA | Chr5:44035468-44035705 |
| Chr5-29 | 缺失 Deletion | -42 | GGAGGAAATTTTGGCTTGGGG | ACCAAAAACATGGAACCACCT | Chr5:44418169-44418332 |
| Chr5-30 | 插入 Insertion | 34 | GCAACTGTCTGATCCTTTGGG | ACTTCGCATTTCAGGGGAGA | Chr5:45207382-45207589 |
| Chr6-09 | 缺失 Deletion | -38 | CTAGACGCCCACGAAGCC | TCTTCCCTCTGCCGACCA | Chr6:9710183-9710427 |
| Chr6-10 | 插入 Insertion | 46 | ACATGCATGCCTAGTCACGA | GGGTTGTTGGAAGGGTCCT | Chr6:10024162-10024384 |
| Chr6-27 | 插入 Insertion | 45 | ATGTTGGGTCGCAGTGGC | AGGGCGGAGGGGATTTCT | Chr6:26274037-26274275 |
| Chr6-28 | 缺失 Deletion | -32 | TGAAAACAACTGCGAAACACT | ACTTCAGATGGGTACTTCTGC | Chr6:26872028-26872241 |
| Chr6-29 | 缺失 Deletion | -44 | CGGCAAGAGGAGAAGGGC | GGGGTTTCCAAGGAAGGGG | Chr6:28571296-28571513 |
| Chr6-32 | 缺失 Deletion | -43 | ACGCTAATGTTGCCGCCA | CGTCGAGCTTTACCCGCA | Chr6:33681522-33681724 |
| Chr7-01 | 插入 Insertion | 37 | TGCTCGGACGACCACAAC | AGTGGCCAACGAGCAGAG | Chr7:12591-12775 |
| Chr7-02 | 缺失 Deletion | -47 | GCAGCGGCCAATGCTAAG | GGGTGGTTGATGCTTCGC | Chr7:30560-30830 |
| Chr7-06 | 插入 Insertion | 31 | GGGGTTTAGCGTGGGTCC | AATGGCTGGTCTCACGCG | Chr7:449549-449763 |
| Chr7-12 | 缺失 Deletion | -46 | TGCATAAGGTCTGGAGACTGT | ACCCGAGCTACACCACAA | Chr7:2509875-2510036 |
| Chr7-19 | 缺失 Deletion | -41 | GCCTAGAGCCTCCAAACCA | GGAGATCCCTGTGCTGCC | Chr7:5959628-5959858 |
| Chr7-21 | 缺失 Deletion | -45 | ACTGAGTGAGCGATGGCAC | TGGGATGGCAGTAAAGCGG | Chr7:6662961-6663207 |
| Chr7-24 | 缺失 Deletion | -49 | ACCCTGGCTAAACGCGTC | ATGGGGGCGGGTCAACTA | Chr7:8693948-8694111 |
| Chr7-28 | 插入 Insertion | 33 | ACTCGTGTTCCCCAAAACA | ACCTATCAATTCTCCCGTCGA | Chr7:11076056-11076250 |
| Chr7-35 | 缺失 Deletion | -38 | AGAACCCCATCAACGGAAACT | ACCTTCCCACCTCACTCGT | Chr7:15440638-15440766 |
| Chr8-09 | 缺失 Deletion | -42 | ATCAGGCAAATGCATGGTTGA | AACCATCCTGGACCCGGA | Chr8:1320235-1320497 |
| Chr8-16 | 缺失 Deletion | -47 | TGGCTCCACAATCTGGCA | TGCCAACTGGTTTCCTCTTTC | Chr8:3251541-3251789 |
| Chr8-18 | 缺失 Deletion | -44 | CCCGCCTTTTCCACGTGA | AGTCATTCTGGTTCGGGTTGA | Chr8:3574827-3575078 |
| Chr8-19 | 缺失 Deletion | -42 | GCTTGCGGCAATTGGTTTC | ACGAGGATTGACAGGCAGC | Chr8:3680573-3680739 |
| Chr8-26 | 缺失 Deletion | -46 | AGTGGCAAGCAGTGTAGGA | CTTTCACTGCCGGGGAGT | Chr8:6135649-6135902 |
| Chr8-29 | 插入 Insertion | 44 | ACATCAGCTGCTCATTCCAT | TTGTACAGTCCCAAGTTGTGT | Chr8:7246002-7246208 |
| Chr8-30 | 缺失 Deletion | -33 | TGAAGGCCCCAGTTTTCCA | TGCATGTATTCGACCTTGTGG | Chr8:7761356-7761536 |
| Chr8-32 | 插入 Insertion | 36 | ACACTGCACTGCACTAGCT | GGCCAGTTTAGCCCGTCT | Chr8:8396708-8396836 |
| Chr9-09 | 缺失 Deletion | -32 | TCCGTTAACGCTCCGCAG | TGTGGTGGTTCTCTCCTTGTG | Chr9:5206000-5206184 |
| Chr9-15 | 插入 Insertion | 45 | CGACATGCAAGGAACTGGG | GACTTGGGTGCCGAAGCT | Chr9:6742034-6742244 |
| Chr9-17 | 缺失 Deletion | -34 | GGCGTTGTATGGCTCACG | GAGATTGCTCAGGTGTGGGA | Chr9:8310123-8310248 |
| Chr9-22 | 插入 Insertion | 30 | CCAACACCCCATTCCCCA | TGCACTAGACTCCTCCATCCA | Chr9:9397358-9397476 |
| Chr9-25 | 缺失 Deletion | -31 | CTTGTCCACGCGTGAGGT | GCTCGTCCCCTTTGCACA | Chr9:12450328-12450471 |
| Chr9-26 | 缺失 Deletion | -39 | CCGGTATCACTGAGGGCC | CAGTCGTGTGGAGTTGGGA | Chr9:14289457-14289653 |
| Chr9-27 | 缺失 Deletion | -31 | CTAGCACCAGGCCGTCAG | CCGCTGCACCTGGAGAAA | Chr9:14871648-14871817 |
| Chr9-31 | 缺失 Deletion | -45 | AGTTAGCCCATGCGCGTT | AGGGCCTCCTCCACTGAC | Chr9:27896823-27896950 |
表2 72对核心InDel标记引物的位置和序列信息
Table 2 Location and sequence information of 72 pairs of core InDel primers
| 标记号 InDel labels | 标记类型 Type | 差异长 度/bp Length | 上游扩增引物(5′-3′) Forward primer | 下游扩增引物(5′-3′) Reverse primer | 标记引物扩增位置 Amplification location |
|---|---|---|---|---|---|
| Chr1-09 | 插入 Insertion | 35 | ACCTTGACATACATGCATGCA | TGCCTTGAGTCACACGGA | Chr1:2749817-2750039 |
| Chr1-10 | 插入 Insertion | 36 | GAACCTCCTCCCCCGCTA | TGAACCACCTGCTCGCAT | Chr1:2857128-2857366 |
| Chr1-15 | 插入 Insertion | 49 | TGTTCCATGTGTTATGGACGT | ACAAGCTTGGTGCATATGCT | Chr1:3213152-3213384 |
| Chr1-26 | 缺失 Deletion | -46 | CGCCCTCTCCCGTAAAGC | ACCACAGGTACTTGGCCG | Chr1:7259239-7259506 |
| Chr1-29 | 缺失 Deletion | -30 | CGTGAGGTGCTCGCTGAT | GGGACATCTCGTGCACGT | Chr1:7846667-7846868 |
| Chr1-30 | 插入 Insertion | 42 | GAACGCAAAAGACTGACTCCT | AGGTTCTGAAGACATGCACTT | Chr1:8015769-8016044 |
| Chr1-39 | 插入 Insertion | 39 | GTCCTCAGGACGACCCTAGA | ATCTGGTGGCAGTGGCAG | Chr1:10631445-10631724 |
| Chr2-01 | 插入 Insertion | 37 | GCTCCAACGAGAGAGATGACT | CGGTGCTGGTGACGAAGA | Chr2:307044-307320 |
| Chr2-02 | 插入 Insertion | 31 | CAGGATTTTGCTAGGGTCAGT | AGACTTGCCTGCTGGAGC | Chr2:612393-612613 |
| Chr2-13 | 缺失 Deletion | -34 | ACCAACCGTCGATCCTCA | TGGCAAAACGTCGTGATGT | Chr2:5215597-5215796 |
| Chr2-15 | 插入 Insertion | 44 | TGCTGGGAATGCTTTGTGC | TGGTCCATGATGCCAGCTG | Chr2:5922591-5922860 |
| Chr2-18 | 插入 Insertion | 44 | GGTTAGACTGTCAAGTGGGCT | CGCCCTGAAAACATGGTGC | Chr2:10719434-10719712 |
| Chr2-20 | 缺失 Deletion | -31 | ACAACCTGAGAGCTCTTCCC | AGCTGCTTCTACCTGCTTGA | Chr2:12083278-12083527 |
| Chr2-29 | 插入 Insertion | 48 | TGGAATCAAGATTTGCACCGA | TTCCAGTTTCCGAACAACCA | Chr2:23526410-23526648 |
| Chr2-31 | 缺失 Deletion | -33 | GGCAGCACTTCATTGCCG | GGGTCTTACACTGGTTGCCA | Chr2:28941802-28942025 |
| Chr2-38 | 插入 Insertion | 38 | ACGAGTCTTCAAGCTTGCCA | TGCCAGTCTGTAAATTGCGG | Chr2:34031300-34031578 |
| Chr3-03 | 插入 Insertion | 48 | GCCAACACAAGAACGGAAACT | TGCAGTTCAAGTGGCGCT | Chr3:627145-627348 |
| Chr3-17 | 缺失 Deletion | -44 | CTCCTGCACAGTCGCCAT | GCTTGCTTTGCTCTCCGC | Chr3:6901713-6901929 |
| Chr3-21 | 缺失 Deletion | -46 | CCCTTCCCCCTCCCAAGA | TTCTGCCGTCGGTTGAGC | Chr3:10826323-10826544 |
| Chr3-22 | 插入 Insertion | 39 | TCCCGGACATCTGACCCC | AGCCTTTGATTCCCACTGCA | Chr3:10886342-10886562 |
| Chr3-23 | 插入 Insertion | 31 | GGAGCACAAGCCATGGGT | AGCCACCAGTCAGACCTCT | Chr3:11351904-11352128 |
| Chr3-30 | 插入 Insertion | 34 | ACGGTGACAGCATGAGGC | CACATGCAAGATGCCGGC | Chr3:16193105-16193338 |
| Chr3-31 | 插入 Insertion | 42 | AGCCAACCCACCACAAGT | CTCAGCCTATTTGTTGGTCCA | Chr3:16707449-16707694 |
| Chr3-33 | 插入 Insertion | 33 | TGCTGGAAAACACCCGCA | GCCTCCCCAAGTCAGCTG | Chr3:21024394-21024593 |
| Chr4-03 | 缺失 Deletion | -33 | CCAGCATCAGAACTCGAGGT | TTGGTTGCGGGGAATGCA | Chr4:1324489-1324712 |
| Chr4-08 | 插入 Insertion | 42 | CGAGCCTGTGTACCTGGT | TCACTTTCGTCAGTTGCCTGA | Chr4:4076944-4077153 |
| Chr4-14 | 插入 Insertion | 41 | GCGGTGAGCCTGAGATGA | AGACTCGTGGAGAACAAAGCA | Chr4:9192243-9192474 |
| Chr4-24 | 缺失 Deletion | -40 | ACGGGGCGTACATGTGTG | ACGAGGCCACCCCTAAGA | Chr4:21621919-21622128 |
| Chr4-25 | 插入 Insertion | 45 | TGGTTTCGCAAGTTCGCA | GTGCTAGTTGTTTTGCATGCT | Chr4:22997264-22997514 |
| Chr4-26 | 缺失 Deletion | -46 | AGAGGGGTTAGGCAGGGG | TCCCCACATTGATGTTGACCA | Chr4:23149465-23149734 |
| Chr4-34 | 缺失 Deletion | -32 | TCACGAGTTGAGGGAGGTCT | ACTTGTTGAGGGTTGAAAGCC | Chr4:28955964-28956194 |
| Chr4-40 | 插入 Insertion | 46 | GACGTGGTGCTTATCCCGT | GGCAGTCCAGTTCTATAGGGG | Chr4:34041449-34041703 |
| Chr5-02 | 缺失 Deletion | -49 | TGCGCACAACATTCAGGTC | GGCTATGTTGTGCCTGGGA | Chr5:59655-59828 |
| Chr5-07 | 插入 Insertion | 38 | TGAGTGGAAAGAAGATGCGGA | TCATCCCTCCATCCACAAACT | Chr5:272642-272800 |
| Chr5-08 | 缺失 Deletion | -34 | GACAAAGCAAAACTGCATCGT | TGATTTGGCAGCCCCTGG | Chr5:699745-699920 |
| Chr5-11 | 缺失 Deletion | -35 | GCGGCAATCAGAGGAGCA | GCACCACACAGACAAAAATCA | Chr5:1024290-1024446 |
| Chr5-15 | 缺失 Deletion | -33 | TGGGAGGTAGCTGACGGA | CAGCATGCCTAACCGTGC | Chr5:1509285-1509412 |
| Chr5-18 | 缺失 Deletion | -33 | GACTCCGATGATGTCCTCTGT | CGTGAAGGAAATCGGTGTGC | Chr5:1615976-1616100 |
| Chr5-24 | 插入 Insertion | 31 | GGGCTTGAGTTTGGGCCT | CCCAGTCCCATACCCCGA | Chr5:44035468-44035705 |
| Chr5-29 | 缺失 Deletion | -42 | GGAGGAAATTTTGGCTTGGGG | ACCAAAAACATGGAACCACCT | Chr5:44418169-44418332 |
| Chr5-30 | 插入 Insertion | 34 | GCAACTGTCTGATCCTTTGGG | ACTTCGCATTTCAGGGGAGA | Chr5:45207382-45207589 |
| Chr6-09 | 缺失 Deletion | -38 | CTAGACGCCCACGAAGCC | TCTTCCCTCTGCCGACCA | Chr6:9710183-9710427 |
| Chr6-10 | 插入 Insertion | 46 | ACATGCATGCCTAGTCACGA | GGGTTGTTGGAAGGGTCCT | Chr6:10024162-10024384 |
| Chr6-27 | 插入 Insertion | 45 | ATGTTGGGTCGCAGTGGC | AGGGCGGAGGGGATTTCT | Chr6:26274037-26274275 |
| Chr6-28 | 缺失 Deletion | -32 | TGAAAACAACTGCGAAACACT | ACTTCAGATGGGTACTTCTGC | Chr6:26872028-26872241 |
| Chr6-29 | 缺失 Deletion | -44 | CGGCAAGAGGAGAAGGGC | GGGGTTTCCAAGGAAGGGG | Chr6:28571296-28571513 |
| Chr6-32 | 缺失 Deletion | -43 | ACGCTAATGTTGCCGCCA | CGTCGAGCTTTACCCGCA | Chr6:33681522-33681724 |
| Chr7-01 | 插入 Insertion | 37 | TGCTCGGACGACCACAAC | AGTGGCCAACGAGCAGAG | Chr7:12591-12775 |
| Chr7-02 | 缺失 Deletion | -47 | GCAGCGGCCAATGCTAAG | GGGTGGTTGATGCTTCGC | Chr7:30560-30830 |
| Chr7-06 | 插入 Insertion | 31 | GGGGTTTAGCGTGGGTCC | AATGGCTGGTCTCACGCG | Chr7:449549-449763 |
| Chr7-12 | 缺失 Deletion | -46 | TGCATAAGGTCTGGAGACTGT | ACCCGAGCTACACCACAA | Chr7:2509875-2510036 |
| Chr7-19 | 缺失 Deletion | -41 | GCCTAGAGCCTCCAAACCA | GGAGATCCCTGTGCTGCC | Chr7:5959628-5959858 |
| Chr7-21 | 缺失 Deletion | -45 | ACTGAGTGAGCGATGGCAC | TGGGATGGCAGTAAAGCGG | Chr7:6662961-6663207 |
| Chr7-24 | 缺失 Deletion | -49 | ACCCTGGCTAAACGCGTC | ATGGGGGCGGGTCAACTA | Chr7:8693948-8694111 |
| Chr7-28 | 插入 Insertion | 33 | ACTCGTGTTCCCCAAAACA | ACCTATCAATTCTCCCGTCGA | Chr7:11076056-11076250 |
| Chr7-35 | 缺失 Deletion | -38 | AGAACCCCATCAACGGAAACT | ACCTTCCCACCTCACTCGT | Chr7:15440638-15440766 |
| Chr8-09 | 缺失 Deletion | -42 | ATCAGGCAAATGCATGGTTGA | AACCATCCTGGACCCGGA | Chr8:1320235-1320497 |
| Chr8-16 | 缺失 Deletion | -47 | TGGCTCCACAATCTGGCA | TGCCAACTGGTTTCCTCTTTC | Chr8:3251541-3251789 |
| Chr8-18 | 缺失 Deletion | -44 | CCCGCCTTTTCCACGTGA | AGTCATTCTGGTTCGGGTTGA | Chr8:3574827-3575078 |
| Chr8-19 | 缺失 Deletion | -42 | GCTTGCGGCAATTGGTTTC | ACGAGGATTGACAGGCAGC | Chr8:3680573-3680739 |
| Chr8-26 | 缺失 Deletion | -46 | AGTGGCAAGCAGTGTAGGA | CTTTCACTGCCGGGGAGT | Chr8:6135649-6135902 |
| Chr8-29 | 插入 Insertion | 44 | ACATCAGCTGCTCATTCCAT | TTGTACAGTCCCAAGTTGTGT | Chr8:7246002-7246208 |
| Chr8-30 | 缺失 Deletion | -33 | TGAAGGCCCCAGTTTTCCA | TGCATGTATTCGACCTTGTGG | Chr8:7761356-7761536 |
| Chr8-32 | 插入 Insertion | 36 | ACACTGCACTGCACTAGCT | GGCCAGTTTAGCCCGTCT | Chr8:8396708-8396836 |
| Chr9-09 | 缺失 Deletion | -32 | TCCGTTAACGCTCCGCAG | TGTGGTGGTTCTCTCCTTGTG | Chr9:5206000-5206184 |
| Chr9-15 | 插入 Insertion | 45 | CGACATGCAAGGAACTGGG | GACTTGGGTGCCGAAGCT | Chr9:6742034-6742244 |
| Chr9-17 | 缺失 Deletion | -34 | GGCGTTGTATGGCTCACG | GAGATTGCTCAGGTGTGGGA | Chr9:8310123-8310248 |
| Chr9-22 | 插入 Insertion | 30 | CCAACACCCCATTCCCCA | TGCACTAGACTCCTCCATCCA | Chr9:9397358-9397476 |
| Chr9-25 | 缺失 Deletion | -31 | CTTGTCCACGCGTGAGGT | GCTCGTCCCCTTTGCACA | Chr9:12450328-12450471 |
| Chr9-26 | 缺失 Deletion | -39 | CCGGTATCACTGAGGGCC | CAGTCGTGTGGAGTTGGGA | Chr9:14289457-14289653 |
| Chr9-27 | 缺失 Deletion | -31 | CTAGCACCAGGCCGTCAG | CCGCTGCACCTGGAGAAA | Chr9:14871648-14871817 |
| Chr9-31 | 缺失 Deletion | -45 | AGTTAGCCCATGCGCGTT | AGGGCCTCCTCCACTGAC | Chr9:27896823-27896950 |
| 标记号 InDel labels | 等位基 因数 (Na) Number of alleles | 有效等位基 因数(Ne) Effective number of alleles | 基因多样性 指数(H) Gene diversity index | 多态性信息 含量(PIC) Polymorphic information content | 遗传距离 (Nei’s) Nei’s genetic distance | 标记号 InDel labels | 等位基 因数 (Na) Number of alleles | 有效等位基 因数(Ne) Effective number of alleles | 基因多样 性指数 (H) Gene diversity index | 多态性信息含量(PIC) Polymorphic information content | 遗传距离 (Nei’s) Nei’s genetic distance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr1-09 | 2.00 | 1.8579 | 0.6544 | 0.3551 | 0.4617 | Chr5-29 | 2.00 | 1.2605 | 0.3609 | 0.1853 | 0.2067 |
| Chr1-10 | 2.00 | 1.9991 | 0.6929 | 0.3749 | 0.4998 | Chr5-30 | 2.00 | 1.2865 | 0.3819 | 0.1979 | 0.2227 |
| Chr1-15 | 2.00 | 1.3665 | 0.4390 | 0.3657 | 0.4817 | Chr6-09 | 2.00 | 1.9919 | 0.6911 | 0.3740 | 0.4980 |
| Chr1-26 | 2.00 | 1.9293 | 0.6747 | 0.2322 | 0.2682 | Chr6-10 | 2.00 | 1.5589 | 0.5441 | 0.2943 | 0.3585 |
| Chr1-29 | 2.00 | 1.1357 | 0.2374 | 0.1124 | 0.1195 | Chr6-27 | 2.00 | 1.3937 | 0.4562 | 0.2426 | 0.2825 |
| Chr1-30 | 2.00 | 1.8776 | 0.6602 | 0.3582 | 0.4674 | Chr6-28 | 2.00 | 1.3937 | 0.4562 | 0.2426 | 0.2825 |
| Chr1-39 | 2.00 | 1.9293 | 0.6747 | 0.3657 | 0.4817 | Chr6-29 | 2.00 | 1.4761 | 0.5033 | 0.2705 | 0.3225 |
| Chr2-01 | 2.00 | 1.9293 | 0.6747 | 0.0957 | 0.1007 | Chr5-30 | 2.00 | 1.2865 | 0.3819 | 0.1979 | 0.2227 |
| Chr2-02 | 2.00 | 2.0000 | 0.6931 | 0.3677 | 0.4855 | Chr6-09 | 2.00 | 1.9919 | 0.6911 | 0.3740 | 0.4980 |
| Chr2-13 | 2.00 | 1.9437 | 0.6786 | 0.3013 | 0.3696 | Chr6-10 | 2.00 | 1.5589 | 0.5441 | 0.2943 | 0.3585 |
| Chr2-15 | 2.00 | 1.6934 | 0.5997 | 0.3657 | 0.4817 | Chr6-27 | 2.00 | 1.3937 | 0.4562 | 0.2426 | 0.2825 |
| Chr2-18 | 2.00 | 1.9856 | 0.6895 | 0.3609 | 0.4726 | Chr6-32 | 2.00 | 1.8579 | 0.6544 | 0.3551 | 0.4617 |
| Chr2-20 | 2.00 | 1.5864 | 0.5564 | 0.3256 | 0.4095 | Chr7-01 | 2.00 | 1.7686 | 0.6262 | 0.2322 | 0.2682 |
| Chr2-29 | 2.00 | 1.1120 | 0.2078 | 0.3732 | 0.4964 | Chr7-02 | 2.00 | 1.8776 | 0.6602 | 0.3013 | 0.3696 |
| Chr2-31 | 2.00 | 2.0000 | 0.6931 | 0.3750 | 0.5000 | Chr7-06 | 2.00 | 1.6136 | 0.5681 | 0.3694 | 0.4889 |
| Chr2-38 | 2.00 | 1.8961 | 0.6655 | 0.3750 | 0.5000 | Chr7-12 | 2.00 | 1.9566 | 0.6820 | 0.3657 | 0.4817 |
| Chr3-03 | 2.00 | 1.1636 | 0.2691 | 0.3709 | 0.4919 | Chr7-19 | 2.00 | 1.7442 | 0.6179 | 0.3402 | 0.4346 |
| Chr3-17 | 2.00 | 1.9293 | 0.6747 | 0.2322 | 0.2682 | Chr7-21 | 2.00 | 1.5864 | 0.5564 | 0.2740 | 0.3277 |
| Chr3-21 | 2.00 | 1.7686 | 0.6262 | 0.3657 | 0.4817 | Chr7-24 | 2.00 | 1.3665 | 0.4390 | 0.3356 | 0.4267 |
| Chr3-22 | 2.00 | 1.9679 | 0.6850 | 0.3677 | 0.4855 | Chr7-28 | 2.00 | 1.9293 | 0.6747 | 0.3582 | 0.4674 |
| Chr3-23 | 2.00 | 1.9437 | 0.6786 | 0.3402 | 0.4346 | Chr7-35 | 2.00 | 1.4875 | 0.5093 | 0.3080 | 0.3803 |
| Chr3-30 | 2.00 | 1.6672 | 0.5897 | 0.3443 | 0.4421 | Chr8-09 | 2.00 | 1.4210 | 0.4727 | 0.0408 | 0.0416 |
| Chr3-31 | 2.00 | 1.7923 | 0.6340 | 0.3201 | 0.4002 | Chr8-16 | 2.00 | 1.0435 | 0.1030 | 0.1853 | 0.2067 |
| Chr3-33 | 2.00 | 1.3665 | 0.4390 | 0.1307 | 0.1406 | Chr8-18 | 2.00 | 1.3665 | 0.4390 | 0.2524 | 0.2963 |
| Chr4-03 | 2.00 | 1.5589 | 0.5441 | 0.3582 | 0.4674 | Chr8-19 | 2.00 | 1.9293 | 0.6747 | 0.3013 | 0.3696 |
| Chr4-08 | 2.00 | 1.5589 | 0.5441 | 0.3551 | 0.4617 | Chr8-26 | 2.00 | 1.5864 | 0.5564 | 0.3740 | 0.4980 |
| Chr4-14 | 2.00 | 1.8579 | 0.6544 | 0.2426 | 0.2825 | Chr8-29 | 2.00 | 1.9293 | 0.6747 | 0.3657 | 0.4817 |
| Chr4-24 | 2.00 | 1.3937 | 0.4562 | 0.1979 | 0.2227 | Chr8-30 | 2.00 | 1.2605 | 0.3609 | 0.2322 | 0.2682 |
| Chr4-25 | 2.00 | 1.8776 | 0.6602 | 0.2943 | 0.3585 | Chr8-32 | 2.00 | 1.9919 | 0.6911 | 0.3657 | 0.4817 |
| Chr4-26 | 2.00 | 1.8078 | 0.6390 | 0.2943 | 0.3585 | Chr9-09 | 3.00 | 2.0664 | 0.8735 | 0.4523 | 0.5161 |
| Chr4-34 | 2.00 | 1.6672 | 0.5897 | 0.3470 | 0.4468 | Chr9-15 | 2.00 | 1.6405 | 0.5792 | 0.3732 | 0.4964 |
| Chr4-40 | 2.00 | 1.2865 | 0.3819 | 0.3201 | 0.4002 | Chr9-17 | 2.00 | 1.5158 | 0.5236 | 0.3634 | 0.4774 |
| Chr5-02 | 2.00 | 1.4210 | 0.4727 | 0.2524 | 0.2963 | Chr9-22 | 2.00 | 1.5589 | 0.5441 | 0.3142 | 0.3904 |
| Chr5-07 | 2.00 | 1.8961 | 0.6655 | 0.3609 | 0.4726 | Chr9-25 | 2.00 | 1.9856 | 0.6895 | 0.2943 | 0.3585 |
| Chr5-08 | 2.00 | 1.6136 | 0.5681 | 0.3080 | 0.3803 | Chr9-26 | 2.00 | 1.9134 | 0.6703 | 0.2868 | 0.3470 |
| Chr5-11 | 2.00 | 1.3129 | 0.4019 | 0.2099 | 0.2383 | Chr9-27 | 2.00 | 1.5314 | 0.5312 | 0.2824 | 0.3403 |
| Chr5-15 | 2.00 | 1.6934 | 0.5997 | 0.3256 | 0.4095 | Chr9-31 | 2.00 | 1.9097 | 0.6693 | 0.3629 | 0.4764 |
| Chr5-18 | 2.00 | 1.2865 | 0.3819 | 0.1979 | 0.2227 | 平均值 | 2.01 | 1.6692 | 0.5660 | 0.3041 | 0.3828 |
| Chr5-24 | 2.00 | 1.9134 | 0.6703 | 0.3634 | 0.4774 | Average |
表 3 47份番木瓜种质的遗传多样性分析
Table 3 Genetic diversity analysis of 47 papaya germplasms
| 标记号 InDel labels | 等位基 因数 (Na) Number of alleles | 有效等位基 因数(Ne) Effective number of alleles | 基因多样性 指数(H) Gene diversity index | 多态性信息 含量(PIC) Polymorphic information content | 遗传距离 (Nei’s) Nei’s genetic distance | 标记号 InDel labels | 等位基 因数 (Na) Number of alleles | 有效等位基 因数(Ne) Effective number of alleles | 基因多样 性指数 (H) Gene diversity index | 多态性信息含量(PIC) Polymorphic information content | 遗传距离 (Nei’s) Nei’s genetic distance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr1-09 | 2.00 | 1.8579 | 0.6544 | 0.3551 | 0.4617 | Chr5-29 | 2.00 | 1.2605 | 0.3609 | 0.1853 | 0.2067 |
| Chr1-10 | 2.00 | 1.9991 | 0.6929 | 0.3749 | 0.4998 | Chr5-30 | 2.00 | 1.2865 | 0.3819 | 0.1979 | 0.2227 |
| Chr1-15 | 2.00 | 1.3665 | 0.4390 | 0.3657 | 0.4817 | Chr6-09 | 2.00 | 1.9919 | 0.6911 | 0.3740 | 0.4980 |
| Chr1-26 | 2.00 | 1.9293 | 0.6747 | 0.2322 | 0.2682 | Chr6-10 | 2.00 | 1.5589 | 0.5441 | 0.2943 | 0.3585 |
| Chr1-29 | 2.00 | 1.1357 | 0.2374 | 0.1124 | 0.1195 | Chr6-27 | 2.00 | 1.3937 | 0.4562 | 0.2426 | 0.2825 |
| Chr1-30 | 2.00 | 1.8776 | 0.6602 | 0.3582 | 0.4674 | Chr6-28 | 2.00 | 1.3937 | 0.4562 | 0.2426 | 0.2825 |
| Chr1-39 | 2.00 | 1.9293 | 0.6747 | 0.3657 | 0.4817 | Chr6-29 | 2.00 | 1.4761 | 0.5033 | 0.2705 | 0.3225 |
| Chr2-01 | 2.00 | 1.9293 | 0.6747 | 0.0957 | 0.1007 | Chr5-30 | 2.00 | 1.2865 | 0.3819 | 0.1979 | 0.2227 |
| Chr2-02 | 2.00 | 2.0000 | 0.6931 | 0.3677 | 0.4855 | Chr6-09 | 2.00 | 1.9919 | 0.6911 | 0.3740 | 0.4980 |
| Chr2-13 | 2.00 | 1.9437 | 0.6786 | 0.3013 | 0.3696 | Chr6-10 | 2.00 | 1.5589 | 0.5441 | 0.2943 | 0.3585 |
| Chr2-15 | 2.00 | 1.6934 | 0.5997 | 0.3657 | 0.4817 | Chr6-27 | 2.00 | 1.3937 | 0.4562 | 0.2426 | 0.2825 |
| Chr2-18 | 2.00 | 1.9856 | 0.6895 | 0.3609 | 0.4726 | Chr6-32 | 2.00 | 1.8579 | 0.6544 | 0.3551 | 0.4617 |
| Chr2-20 | 2.00 | 1.5864 | 0.5564 | 0.3256 | 0.4095 | Chr7-01 | 2.00 | 1.7686 | 0.6262 | 0.2322 | 0.2682 |
| Chr2-29 | 2.00 | 1.1120 | 0.2078 | 0.3732 | 0.4964 | Chr7-02 | 2.00 | 1.8776 | 0.6602 | 0.3013 | 0.3696 |
| Chr2-31 | 2.00 | 2.0000 | 0.6931 | 0.3750 | 0.5000 | Chr7-06 | 2.00 | 1.6136 | 0.5681 | 0.3694 | 0.4889 |
| Chr2-38 | 2.00 | 1.8961 | 0.6655 | 0.3750 | 0.5000 | Chr7-12 | 2.00 | 1.9566 | 0.6820 | 0.3657 | 0.4817 |
| Chr3-03 | 2.00 | 1.1636 | 0.2691 | 0.3709 | 0.4919 | Chr7-19 | 2.00 | 1.7442 | 0.6179 | 0.3402 | 0.4346 |
| Chr3-17 | 2.00 | 1.9293 | 0.6747 | 0.2322 | 0.2682 | Chr7-21 | 2.00 | 1.5864 | 0.5564 | 0.2740 | 0.3277 |
| Chr3-21 | 2.00 | 1.7686 | 0.6262 | 0.3657 | 0.4817 | Chr7-24 | 2.00 | 1.3665 | 0.4390 | 0.3356 | 0.4267 |
| Chr3-22 | 2.00 | 1.9679 | 0.6850 | 0.3677 | 0.4855 | Chr7-28 | 2.00 | 1.9293 | 0.6747 | 0.3582 | 0.4674 |
| Chr3-23 | 2.00 | 1.9437 | 0.6786 | 0.3402 | 0.4346 | Chr7-35 | 2.00 | 1.4875 | 0.5093 | 0.3080 | 0.3803 |
| Chr3-30 | 2.00 | 1.6672 | 0.5897 | 0.3443 | 0.4421 | Chr8-09 | 2.00 | 1.4210 | 0.4727 | 0.0408 | 0.0416 |
| Chr3-31 | 2.00 | 1.7923 | 0.6340 | 0.3201 | 0.4002 | Chr8-16 | 2.00 | 1.0435 | 0.1030 | 0.1853 | 0.2067 |
| Chr3-33 | 2.00 | 1.3665 | 0.4390 | 0.1307 | 0.1406 | Chr8-18 | 2.00 | 1.3665 | 0.4390 | 0.2524 | 0.2963 |
| Chr4-03 | 2.00 | 1.5589 | 0.5441 | 0.3582 | 0.4674 | Chr8-19 | 2.00 | 1.9293 | 0.6747 | 0.3013 | 0.3696 |
| Chr4-08 | 2.00 | 1.5589 | 0.5441 | 0.3551 | 0.4617 | Chr8-26 | 2.00 | 1.5864 | 0.5564 | 0.3740 | 0.4980 |
| Chr4-14 | 2.00 | 1.8579 | 0.6544 | 0.2426 | 0.2825 | Chr8-29 | 2.00 | 1.9293 | 0.6747 | 0.3657 | 0.4817 |
| Chr4-24 | 2.00 | 1.3937 | 0.4562 | 0.1979 | 0.2227 | Chr8-30 | 2.00 | 1.2605 | 0.3609 | 0.2322 | 0.2682 |
| Chr4-25 | 2.00 | 1.8776 | 0.6602 | 0.2943 | 0.3585 | Chr8-32 | 2.00 | 1.9919 | 0.6911 | 0.3657 | 0.4817 |
| Chr4-26 | 2.00 | 1.8078 | 0.6390 | 0.2943 | 0.3585 | Chr9-09 | 3.00 | 2.0664 | 0.8735 | 0.4523 | 0.5161 |
| Chr4-34 | 2.00 | 1.6672 | 0.5897 | 0.3470 | 0.4468 | Chr9-15 | 2.00 | 1.6405 | 0.5792 | 0.3732 | 0.4964 |
| Chr4-40 | 2.00 | 1.2865 | 0.3819 | 0.3201 | 0.4002 | Chr9-17 | 2.00 | 1.5158 | 0.5236 | 0.3634 | 0.4774 |
| Chr5-02 | 2.00 | 1.4210 | 0.4727 | 0.2524 | 0.2963 | Chr9-22 | 2.00 | 1.5589 | 0.5441 | 0.3142 | 0.3904 |
| Chr5-07 | 2.00 | 1.8961 | 0.6655 | 0.3609 | 0.4726 | Chr9-25 | 2.00 | 1.9856 | 0.6895 | 0.2943 | 0.3585 |
| Chr5-08 | 2.00 | 1.6136 | 0.5681 | 0.3080 | 0.3803 | Chr9-26 | 2.00 | 1.9134 | 0.6703 | 0.2868 | 0.3470 |
| Chr5-11 | 2.00 | 1.3129 | 0.4019 | 0.2099 | 0.2383 | Chr9-27 | 2.00 | 1.5314 | 0.5312 | 0.2824 | 0.3403 |
| Chr5-15 | 2.00 | 1.6934 | 0.5997 | 0.3256 | 0.4095 | Chr9-31 | 2.00 | 1.9097 | 0.6693 | 0.3629 | 0.4764 |
| Chr5-18 | 2.00 | 1.2865 | 0.3819 | 0.1979 | 0.2227 | 平均值 | 2.01 | 1.6692 | 0.5660 | 0.3041 | 0.3828 |
| Chr5-24 | 2.00 | 1.9134 | 0.6703 | 0.3634 | 0.4774 | Average |

图3 基于InDel分子标记的47份番木瓜种质聚类分析图 外圈热图由内至外分别对应花色(白花与黄花分别赋值0和1)、单果质量[0、1、2依次代表轻果(单果质量 < 800 g)、中果(800 g < 单果质量 < 1 200 g)、重果(单果质量 > 1 200 g)]、果形(0、1、2、3、4依次表示梨形、圆形、椭圆形、长椭圆形和长条形)、果肉颜色(0、1、2、3依次表示黄色、橙黄色、橙红色、红色)
Fig. 3 Cluster analysis of 47 papaya varieties /germplasm resources based on InDel molecular marker analysis The outer ring heatmap,from the inside out,corresponds to flower color(white flowers and yellow flowers are assigned 0 and 1,respectively),single fruit weight[0,1,and 2 represent light fruits(single fruit weight < 800 g),medium fruits(800 g ≤ single fruit weight ≤ 1 200 g),and heavy fruits(single fruit weight > 1 200 g)in sequence],fruit shape(0,1,2,3,and 4 represent pyriform,round,elliptical,long elliptical,and enlongated shape in sequence),and flesh color(0,1,2,and 3 represent yellow,orange-yellow,orange-red,and red in sequence)
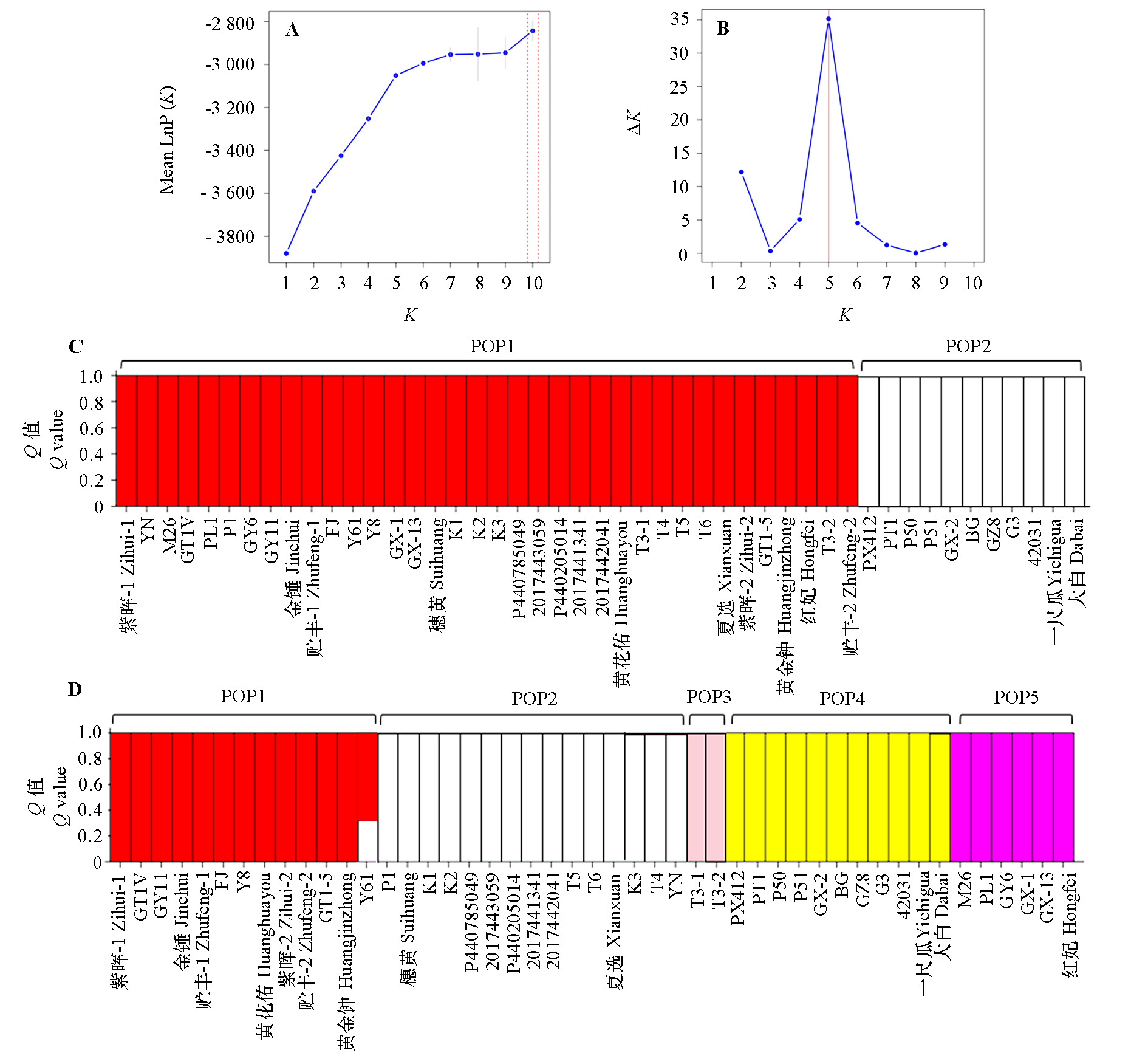
图4 基于InDel分子标记的47份番木瓜种质的群体结构分析 A:lnP(K)值与K值变化折线图;B:Δ K值随K值变化折线图;C:K = 2时的群体结构;D:K = 5时的群体结构
Fig. 4 Population structure analysis of 47 papaya germplasms based on InDel molecular markers A:Line plot of lnP(K)versus K value;B:Line plot of ΔK value versus K value;C:Population structure at K = 2;D:Population structure at K = 5
| 编号 No. | 种质 Germplasm | 花色 Flower color | 单果质量 Fruit weight | 果实形状 Fruit shape | 果肉颜色 Pulp color | 分子身份证编码 Molecular ID |
|---|---|---|---|---|---|---|
| 1 | PX412 | 黄Yellow | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 11221101011001101010111001011010100110100111 |
| 2 | 紫晖-1 Zihui-1 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11320111010101101111111010101111111101011011 |
| 3 | YN | 白White | 轻Light | 椭圆Ellipse | 黄Yellow | 00200101111101110110111111100111100111111001 |
| 4 | M26 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221110101110110111111111011111011011111111 |
| 5 | GT1V | 黄Yellow | 重Heavy | 长椭圆Long ellipse | 橙红Orange-red | 12320110110101111010101110100111100101111010 |
| 6 | PL1 | 白White | 轻Light | 椭圆Ellipse | 橙红Orange-red | 00221110111110100101111111011001110110100101 |
| 7 | PT1 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321110011011100110101001011111010110100101 |
| 8 | P1 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01320111010101010101101011011010110110111001 |
| 9 | P50 | 白White | 轻Light | 长椭圆Long ellipse | 橙红Orange-red | 00321110111011110110100111010111101101110110 |
| 10 | P51 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311001011011111010100111011111100101100101 |
| 11 | GY6 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01310110101110100111111011011001011001101010 |
| 12 | GY11 | 白White | 重Heavy | 长椭圆Long ellipse | 橙红Orange-red | 02321111111111011101101110101110110111111110 |
| 13 | 金锤Jinchui | 黄Yellow | 轻Light | 梨形Pear shaped | 橙红Orange-red | 10021010110101111101101010111110011111111010 |
| 14 | 贮丰-1 Zhufeng-1 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221111011010111101100111010111011011011110 |
| 15 | FJ | 白White | 轻Light | 长椭圆Long ellipse | 黄Yellow | 00301111110101011001111010100111010101111010 |
| 16 | Y61 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01320110111110010101101010101011100111111010 |
| 17 | Y8 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01310110110101011010101010100111100101101010 |
| 18 | GX-1 | 白White | 轻Light | 椭圆Ellipse | 橙红Orange-red | 00221110110110010111101001011111011001100101 |
| 19 | GX-2 | 白White | 轻Light | 椭圆Ellipse | 红Red | 00230110111001101110100101011111100111100101 |
| 20 | GX-13 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311110110110010111011001011011010111111010 |
| 21 | 穗黄Suihuang | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311101111111111011011110010110110111100110 |
| 22 | BG | 白White | 轻Light | 梨形Pear shaped | 橙红Orange-red | 00020110101010100110100101010110010101100101 |
| 23 | K1 | 白White | 中Moderate | 圆形Round | 橙黄Orange-yellow | 01111011110101010110111011011010100110101001 |
| 24 | K2 | 黄Yellow | 轻Light | 长椭圆Long ellipse | 黄Yellow | 10301011011111011011101011011110100111110111 |
| 25 | K3 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321111111111110111110110011101111011111110 |
| 26 | P440785049 | 白White | 中Moderate | 椭圆Ellipse | 橙黄Orange-yellow | 01211010100111110101010110011011111010111110 |
| 27 | 2017443059 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311011101101100111010111101011110111101110 |
| 28 | P440205014 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321010111101101101011011101010101010100110 |
| 29 | GZ8 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311111111010101110101101011110110110100101 |
| 30 | 2017441341 | 白White | 中Moderate | 圆形Round | 橙红Orange-red | 01120111111111010111101011011110100110111101 |
| 31 | 2017442041 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221010110101110111011011011011101011111111 |
| 32 | 黄花佑 Huanghuayou | 黄Yellow | 中Moderate | 梨形 Pear shaped | 橙红Orange-red | 11020111111111011011101010100110101011111111 |
| 33 | G3 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321010011010100110100111100110111101011010 |
| 34 | T3-1 | 白White | 中Moderate | 长椭圆Long ellipse | 黄Yellow | 01301001010110101010011010101010101001011010 |
| 35 | T4 | 白White | 重Heavy | 长椭圆Long ellipse | 橙黄Orange-yellow | 02310111111111110111110110111101111011111110 |
| 36 | T5 | 白White | 重Heavy | 长条形Long strip | 橙黄Orange-yellow | 02411010100110011010100110101011101010011010 |
| 37 | T6 | 白White | 中Moderate | 长条形Long strip | 橙黄Orange-yellow | 01411001011001011010011001011011101010101010 |
| 38 | 夏选Xiaxuan | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01220101011111111110011111011111111011100111 |
| 39 | 42031 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11321001011010011010101010010111100110100101 |
| 40 | 一尺瓜 Yichigua | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221001011010100110101001011111010101100101 |
| 41 | 紫晖-2 Zihui-2 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11320111010101101111111010101111111101011011 |
| 42 | GT1-5 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11320111010101101111111010101111111101011011 |
| 43 | 大白Dabai | 黄Yellow | 轻Light | 椭圆Ellipse | 橙红Orange-red | 10221111011011111010101111011110100111100101 |
| 44 | 黄金钟 Huangjin- zhong | 黄Yellow | 中Moderate | 圆形Round | 橙红Orange-red | 11121111110101011001111010110110010101111010 |
| 45 | 红妃Hongfei | 黄Yellow | 重Heavy | 长椭圆Long ellipse | 橙红Orange-red | 12321010100110010101011110101001010110011010 |
| 46 | T3-2 | 白White | 中Moderate | 长椭圆Long ellipse | 黄Yellow | 01301001100110101010011010101010101001011010 |
| 47 | 贮丰-2 Zhufeng-2 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221111011010111101100111010110011011011110 |
表4 47份番木瓜种质的表型与基因型编码
Table 4 Phenotypic and genotypic codes of 47 papaya germplasms
| 编号 No. | 种质 Germplasm | 花色 Flower color | 单果质量 Fruit weight | 果实形状 Fruit shape | 果肉颜色 Pulp color | 分子身份证编码 Molecular ID |
|---|---|---|---|---|---|---|
| 1 | PX412 | 黄Yellow | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 11221101011001101010111001011010100110100111 |
| 2 | 紫晖-1 Zihui-1 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11320111010101101111111010101111111101011011 |
| 3 | YN | 白White | 轻Light | 椭圆Ellipse | 黄Yellow | 00200101111101110110111111100111100111111001 |
| 4 | M26 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221110101110110111111111011111011011111111 |
| 5 | GT1V | 黄Yellow | 重Heavy | 长椭圆Long ellipse | 橙红Orange-red | 12320110110101111010101110100111100101111010 |
| 6 | PL1 | 白White | 轻Light | 椭圆Ellipse | 橙红Orange-red | 00221110111110100101111111011001110110100101 |
| 7 | PT1 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321110011011100110101001011111010110100101 |
| 8 | P1 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01320111010101010101101011011010110110111001 |
| 9 | P50 | 白White | 轻Light | 长椭圆Long ellipse | 橙红Orange-red | 00321110111011110110100111010111101101110110 |
| 10 | P51 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311001011011111010100111011111100101100101 |
| 11 | GY6 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01310110101110100111111011011001011001101010 |
| 12 | GY11 | 白White | 重Heavy | 长椭圆Long ellipse | 橙红Orange-red | 02321111111111011101101110101110110111111110 |
| 13 | 金锤Jinchui | 黄Yellow | 轻Light | 梨形Pear shaped | 橙红Orange-red | 10021010110101111101101010111110011111111010 |
| 14 | 贮丰-1 Zhufeng-1 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221111011010111101100111010111011011011110 |
| 15 | FJ | 白White | 轻Light | 长椭圆Long ellipse | 黄Yellow | 00301111110101011001111010100111010101111010 |
| 16 | Y61 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01320110111110010101101010101011100111111010 |
| 17 | Y8 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01310110110101011010101010100111100101101010 |
| 18 | GX-1 | 白White | 轻Light | 椭圆Ellipse | 橙红Orange-red | 00221110110110010111101001011111011001100101 |
| 19 | GX-2 | 白White | 轻Light | 椭圆Ellipse | 红Red | 00230110111001101110100101011111100111100101 |
| 20 | GX-13 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311110110110010111011001011011010111111010 |
| 21 | 穗黄Suihuang | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311101111111111011011110010110110111100110 |
| 22 | BG | 白White | 轻Light | 梨形Pear shaped | 橙红Orange-red | 00020110101010100110100101010110010101100101 |
| 23 | K1 | 白White | 中Moderate | 圆形Round | 橙黄Orange-yellow | 01111011110101010110111011011010100110101001 |
| 24 | K2 | 黄Yellow | 轻Light | 长椭圆Long ellipse | 黄Yellow | 10301011011111011011101011011110100111110111 |
| 25 | K3 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321111111111110111110110011101111011111110 |
| 26 | P440785049 | 白White | 中Moderate | 椭圆Ellipse | 橙黄Orange-yellow | 01211010100111110101010110011011111010111110 |
| 27 | 2017443059 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311011101101100111010111101011110111101110 |
| 28 | P440205014 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321010111101101101011011101010101010100110 |
| 29 | GZ8 | 白White | 中Moderate | 长椭圆Long ellipse | 橙黄Orange-yellow | 01311111111010101110101101011110110110100101 |
| 30 | 2017441341 | 白White | 中Moderate | 圆形Round | 橙红Orange-red | 01120111111111010111101011011110100110111101 |
| 31 | 2017442041 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221010110101110111011011011011101011111111 |
| 32 | 黄花佑 Huanghuayou | 黄Yellow | 中Moderate | 梨形 Pear shaped | 橙红Orange-red | 11020111111111011011101010100110101011111111 |
| 33 | G3 | 白White | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 01321010011010100110100111100110111101011010 |
| 34 | T3-1 | 白White | 中Moderate | 长椭圆Long ellipse | 黄Yellow | 01301001010110101010011010101010101001011010 |
| 35 | T4 | 白White | 重Heavy | 长椭圆Long ellipse | 橙黄Orange-yellow | 02310111111111110111110110111101111011111110 |
| 36 | T5 | 白White | 重Heavy | 长条形Long strip | 橙黄Orange-yellow | 02411010100110011010100110101011101010011010 |
| 37 | T6 | 白White | 中Moderate | 长条形Long strip | 橙黄Orange-yellow | 01411001011001011010011001011011101010101010 |
| 38 | 夏选Xiaxuan | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01220101011111111110011111011111111011100111 |
| 39 | 42031 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11321001011010011010101010010111100110100101 |
| 40 | 一尺瓜 Yichigua | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221001011010100110101001011111010101100101 |
| 41 | 紫晖-2 Zihui-2 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11320111010101101111111010101111111101011011 |
| 42 | GT1-5 | 黄Yellow | 中Moderate | 长椭圆Long ellipse | 橙红Orange-red | 11320111010101101111111010101111111101011011 |
| 43 | 大白Dabai | 黄Yellow | 轻Light | 椭圆Ellipse | 橙红Orange-red | 10221111011011111010101111011110100111100101 |
| 44 | 黄金钟 Huangjin- zhong | 黄Yellow | 中Moderate | 圆形Round | 橙红Orange-red | 11121111110101011001111010110110010101111010 |
| 45 | 红妃Hongfei | 黄Yellow | 重Heavy | 长椭圆Long ellipse | 橙红Orange-red | 12321010100110010101011110101001010110011010 |
| 46 | T3-2 | 白White | 中Moderate | 长椭圆Long ellipse | 黄Yellow | 01301001100110101010011010101010101001011010 |
| 47 | 贮丰-2 Zhufeng-2 | 白White | 中Moderate | 椭圆Ellipse | 橙红Orange-red | 01221111011010111101100111010110011011011110 |
| [1] |
doi: 10.1139/g09-043 pmid: 19767901 |
| [2] |
doi: 10.1038/s41598-020-79401-z pmid: 33431939 |
| [3] |
pmid: 6247908 |
| [4] |
|
| [5] |
|
| [6] |
doi: 10.16420/j.issn.0513-353x.2024-0066 |
|
陈方策, 胡平正, 解为玮, 王钲彭, 钟创南, 李海炎, 何业华, 彭泽, 万保雄, 刘朝阳. 2024, 基于重测序的中国李基因组 InDel 标记的开发及应用. 园艺学报, 51 (11):2510-2522.
doi: 10.16420/j.issn.0513-353x.2024-0066 |
|
| [7] |
doi: 10.4238/2015.March.30.8 pmid: 25867396 |
| [8] |
|
| [9] |
|
| [10] |
doi: 10.1111/j.1744-7909.2012.01115.x |
| [11] |
|
| [12] |
|
|
胡陶铸, 林丽, 胡继军, 邰连赛, 王圣洁, 刘杨. 2019. 利用InDel标记构建番茄新品种指纹图谱. 上海交通大学学报(农业科学版), 37 (2):24-29.
|
|
| [13] |
|
| [14] |
pmid: 12033619 |
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
doi: 10.1534/genetics.166.1.419 pmid: 15020433 |
| [19] |
|
| [20] |
doi: 10.1186/s12870-019-2043-0 pmid: 31655544 |
| [21] |
doi: 10.1270/jsbbs.21021 pmid: 35087324 |
| [22] |
NYT 2519-2013. Guidelines for the testing of distinctness,uniformity and stability —Papaya(Carica papaya L.) (in Chinese)
|
|
NYT 2519-2013. 植物新品种特异性、一致性和稳定性测试指南番木瓜
|
|
| [23] |
|
| [24] |
|
| [25] |
doi: 10.16420/j.issn.0513-353x.2021-0935 URL |
|
汤雨晴, 杨惠栋, 闫承璞, 王斯妤, 王雨亭, 胡钟东, 朱方红. 2023. 基于重测序的‘金兰柚’基因组InDel标记的开发及应用. 园艺学报, 50 (1):15-26.
doi: 10.16420/j.issn.0513-353x.2021-0935 URL |
|
| [26] |
|
|
仝征, 彭存智, 常丽丽, 王丹, 黄超, 徐兵强. 2024. 一种高浓度琼脂糖电泳凝胶的制备方法及其应用:中国,CN114397350A. 2024-04-12.
|
|
| [27] |
pmid: 12582698 |
| [28] |
doi: 10.1101/gr.183905.114 pmid: 25762551 |
| [29] |
|
| [30] |
|
|
王珏. 2017. 基于中国樱桃全基因组开发的InDel标记及引物通用性评价[硕士论文]. 雅安: 四川农业大学.
|
|
| [31] |
|
| [32] |
doi: 10.3724/SP.J.1006.2019.84100 |
|
吴迷, 汪念, 沈超, 黄聪, 温天旺, 林忠旭. 2019. 基于重测序的陆地棉InDel标记开发与评价. 作物学报, 45 (2):196-203.
doi: 10.3724/SP.J.1006.2019.84100 |
|
| [33] |
|
| [34] |
|
|
杨敏, 周陈平, 李庆萌, 邝瑞彬, 吴夏明, 魏岳荣. 2024. ‘黄花佑’和‘金锤’番木瓜贮藏期品质、贮藏特性及转录组分析. 中国农学通报, 40 (6):57-66.
doi: 10.11924/j.issn.1000-6850.casb2023-0118 |
|
| [35] |
|
|
杨易, 黎庭耀, 李国景, 陈汉才, 沈卓, 周轩, 吴增祥, 吴新义, 张艳. 2022. 基于重测序的长豇豆基因组InDel标记开发及应用. 园艺学报, 49 (4):778-790.
doi: 10.16420/j.issn.0513-353x.2021-0347 URL |
|
| [36] |
|
|
尤园园, 王帅, 方莹莹, 齐诚, 李淑培, 张映, 陈钰辉, 刘伟, 刘富中, 舒金帅. 2024. 茄子全基因组InDel变异特征及分子标记开发和应用. 园艺学报, 51 (3):520-532.
|
|
| [37] |
doi: 10.1038/s41588-022-01068-1 pmid: 35551309 |
| [38] |
|
|
张国儒, 唐亚萍, 杨涛, 帕提古丽, 王柏柯, 李宁, 王娟, 李晓琴, 余庆辉, 杨生保. 2019. 基于重测序番茄InDel标记的开发. 分子植物育种, 17 (14):4692-4697.
|
|
| [39] |
|
| [40] |
|
|
赵光远. 2020. 番木瓜畸形花叶病毒在寄主基因功能鉴定及交叉保护中的应用[博士论文]. 海口: 海南大学.
|
|
| [41] |
|
|
张颖聪, 任鹏荣, 周常清, 罗金棠, 陈健. 2020. 基于SRAP标记的番木瓜种质资源遗传多样性及亲缘关系分析初报. 中国热带农业,(1):52-59.
|
| [1] | 石兰蓉, 郑剑, 张继, 欧良华, 甘卫堂. 加工型番木瓜新品种‘桂热3号’[J]. 园艺学报, 2025, 52(S2): 83-84. |
| [2] | 钟声远, 罗宇婷, 陈剑锋, 钟海丰, 陈宇华, 刘中华. 蝴蝶兰品种DUS测试数量性状分级及遗传多样性研究[J]. 园艺学报, 2025, 52(7): 1817-1827. |
| [3] | 李佐, 肖文芳, 陈和明, 吕复兵. 蝴蝶兰种质资源遗传多样性及核心种质构建[J]. 园艺学报, 2025, 52(6): 1519-1529. |
| [4] | 孙佩, 张宏, 杨媛, 王华, 李茂福, 康岩慧, 孙向一, 金万梅. 基于SSR标记的蔷薇属种质资源遗传多样性分析及指纹图谱构建[J]. 园艺学报, 2025, 52(6): 1539-1552. |
| [5] | 毕清芮, 崔东升, 马新院, 薛玉冉, 章世奎, 樊国全, 牛莹莹. 新疆本地梨品种的遗传多样性及亲缘关系[J]. 园艺学报, 2025, 52(3): 561-574. |
| [6] | 罗嗣芳, 严翔, 谢丽芳, 孙静贤, 郭紫晶, 张祖铭, 陈兆星. 赣南野生山金柑全基因组InDel标记开发及应用[J]. 园艺学报, 2025, 52(2): 267-278. |
| [7] | 张露, 张平萍, 解玮佳, 许凤, 彭绿春, 宋杰, 杜光辉, 李世峰. 常绿杜鹃种质遗传多样性分析及SSR指纹图谱构建[J]. 园艺学报, 2025, 52(2): 380-394. |
| [8] | 肖熙鸥, 聂珩, 林文秋, 吴彩玉. 茄子全基因组SNP标记的开发[J]. 园艺学报, 2025, 52(1): 88-100. |
| [9] | 秦子璐, 徐正康, 戴晓港, 陈赢男. 望春玉兰种质资源遗传多样性分析与核心种质构建[J]. 园艺学报, 2024, 51(8): 1823-1832. |
| [10] | 秦宇, 郭荣琨, 农惠兰, 王燕, 崔凯, 董宁光. 利用SSR荧光标记构建山楂种质分子身份证[J]. 园艺学报, 2024, 51(6): 1227-1240. |
| [11] | 袁娜, 徐勤圆, 徐照龙, 周玲, 刘晓庆, 陈新, 杜建厂. 基于靶向测序技术的菜豆SNP液相芯片开发及验证[J]. 园艺学报, 2024, 51(5): 1017-1032. |
| [12] | 游平, 杨进, 周俊, 黄爱军, 鲍敏丽, 易龙. 柑橘黄龙病菌原噬菌体的遗传多样性分析[J]. 园艺学报, 2024, 51(4): 727-736. |
| [13] | 尤园园, 王帅, 方莹莹, 齐诚, 李淑培, 张映, 陈钰辉, 刘伟, 刘富中, 舒金帅. 茄子全基因组InDel变异特征及分子标记开发和应用[J]. 园艺学报, 2024, 51(3): 520-532. |
| [14] | 陈方策, 胡平正, 解为玮, 王钲彭, 钟创南, 李海炎, 何业华, 彭泽, 万保雄, 刘朝阳. 基于重测序的中国李基因组InDel标记的开发及应用[J]. 园艺学报, 2024, 51(11): 2510-2522. |
| [15] | 王荣波, 王海云, 张前荣, 蔡松龄, 李本金, 张美祥, 刘裴清. 福建省番茄青枯病菌遗传多样性分析及其抗性砧木的筛选[J]. 园艺学报, 2024, 51(11): 2540-2554. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司