
Acta Horticulturae Sinica ›› 2022, Vol. 49 ›› Issue (1): 129-140.doi: 10.16420/j.issn.0513-353x.2020-1033
• Research Notes • Previous Articles Next Articles
YU Lin1,2, SHE Xiaoman1, TANG Yafei1, LAN Guobing1, LI Zhenggang1, HE Zifu1,2,*( )
)
Received:2021-07-18
Revised:2021-08-19
Online:2022-01-25
Published:2022-01-24
Contact:
HE Zifu
E-mail:hezf@gdppri.com
CLC Number:
YU Lin, SHE Xiaoman, TANG Yafei, LAN Guobing, LI Zhenggang, HE Zifu. Identification of Pathogen Causing Allium ascalonicum Anthracnose in Guangdong[J]. Acta Horticulturae Sinica, 2022, 49(1): 129-140.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-1033
| 目的序列 Target sequence | 引物名称 Primer | 引物序列(5′-3′) Primer sequence | 参考文献 Reference |
|---|---|---|---|
| ITS | ITS1F | CTTGGTCATTTAGAGGAAGTAA | Gardes & Bruns, |
| ITS4 | TCCTCCGCTTATTGATATGC | White et al., | |
| ACT | ACT-512F | ATGTGCAAGGCCGGTTTCGC | Carbone & Kohn, |
| ACT-783R | TACGAGTCCTTCTGGCCCAT | Carbone & Kohn, | |
| CHS1 | CHS-79F | TGGGGCAAGGATGCTTGGAAGAAG | Carbone & Kohn, |
| CHS-354R | TGGAAGAACCATCTGTGAGAGTTG | Carbone & Kohn, | |
| GAPDH | GDF1 | GCCGTCAACGACCCCTTCATT | Guerber et al., |
| GDR1 | GGGTGGAGTCGTACTTGAGCATGT | Guerber et al., | |
| TUB2 | T1 | AACATGCGTGAGATTGTAAGT | O’ Donnell & Cigelnik, |
| Btub4R | CCRGAYTGRCCRAARACRAAGTTGTC | Aveskamp et al., |
Table 1 PCR Primers for ITS,ACT,CHS1,GAPDH and TUB2 genes amplification
| 目的序列 Target sequence | 引物名称 Primer | 引物序列(5′-3′) Primer sequence | 参考文献 Reference |
|---|---|---|---|
| ITS | ITS1F | CTTGGTCATTTAGAGGAAGTAA | Gardes & Bruns, |
| ITS4 | TCCTCCGCTTATTGATATGC | White et al., | |
| ACT | ACT-512F | ATGTGCAAGGCCGGTTTCGC | Carbone & Kohn, |
| ACT-783R | TACGAGTCCTTCTGGCCCAT | Carbone & Kohn, | |
| CHS1 | CHS-79F | TGGGGCAAGGATGCTTGGAAGAAG | Carbone & Kohn, |
| CHS-354R | TGGAAGAACCATCTGTGAGAGTTG | Carbone & Kohn, | |
| GAPDH | GDF1 | GCCGTCAACGACCCCTTCATT | Guerber et al., |
| GDR1 | GGGTGGAGTCGTACTTGAGCATGT | Guerber et al., | |
| TUB2 | T1 | AACATGCGTGAGATTGTAAGT | O’ Donnell & Cigelnik, |
| Btub4R | CCRGAYTGRCCRAARACRAAGTTGTC | Aveskamp et al., |
| 种类 | 菌株 | GenBank登录号GenBank accession number | ||||
|---|---|---|---|---|---|---|
| Species | Isolate | ITS | ACT | CHS1 | GAPDH | TUB2 |
| Colletotrichum lilii | CBS 186.30 | GU227811 | GU227909 | GU228301 | GU228203 | GU228105 |
| CBS 109214 | GU227810 | GU227908 | GU228300 | GU228202 | GU228104 | |
| C. liriopes | CBS 119444 | GU227804 | GU227902 | GU228294 | GU228196 | GU228098 |
| C. spaethianum | BLC1 | MW295524 | MW298835 | MW298836 | MW298837 | MW298838 |
| CBS 167.49 | GU227807 | GU227905 | GU228297 | GU228199 | GU228101 | |
| CBS 100063 | GU227808 | GU227906 | GU228298 | GU228200 | GU228102 | |
| CBS 101631 | GU227809 | GU227907 | GU228299 | GU228201 | GU228103 | |
| WYC1 | MW295525 | MW298839 | MW298840 | MW298841 | MW298842 | |
| C. tofieldiae | CBS 495.85 | GU227801 | GU227899 | GU228291 | GU228193 | GU228095 |
| C. trichellum | CBS 217.64 | GU227812 | GU227910 | GU228302 | GU228204 | GU228106 |
| C. verruculosum | IMI 45525 | GU227806 | GU227904 | GU228296 | GU228198 | GU228100 |
| C. gloeosporioides | CBS 112999 | JX010152 | JX009531 | JX009818 | JX010056 | JX010445 |
| C. theobromicola | ICMP 18649 | JX010294 | JX009444 | JX009869 | JX010006 | JX010447 |
| C. truncatum | CBS 151.35 | GU227862 | GU227960 | GU228352 | GU228254 | GU228156 |
| Monilochaetes infuscans | CBS 869.96 | JQ005780 | JQ005843 | JQ005801 | JX546612 | JQ005864 |
Table 2 Reference isolates used in this study and their GenBank accession numbers
| 种类 | 菌株 | GenBank登录号GenBank accession number | ||||
|---|---|---|---|---|---|---|
| Species | Isolate | ITS | ACT | CHS1 | GAPDH | TUB2 |
| Colletotrichum lilii | CBS 186.30 | GU227811 | GU227909 | GU228301 | GU228203 | GU228105 |
| CBS 109214 | GU227810 | GU227908 | GU228300 | GU228202 | GU228104 | |
| C. liriopes | CBS 119444 | GU227804 | GU227902 | GU228294 | GU228196 | GU228098 |
| C. spaethianum | BLC1 | MW295524 | MW298835 | MW298836 | MW298837 | MW298838 |
| CBS 167.49 | GU227807 | GU227905 | GU228297 | GU228199 | GU228101 | |
| CBS 100063 | GU227808 | GU227906 | GU228298 | GU228200 | GU228102 | |
| CBS 101631 | GU227809 | GU227907 | GU228299 | GU228201 | GU228103 | |
| WYC1 | MW295525 | MW298839 | MW298840 | MW298841 | MW298842 | |
| C. tofieldiae | CBS 495.85 | GU227801 | GU227899 | GU228291 | GU228193 | GU228095 |
| C. trichellum | CBS 217.64 | GU227812 | GU227910 | GU228302 | GU228204 | GU228106 |
| C. verruculosum | IMI 45525 | GU227806 | GU227904 | GU228296 | GU228198 | GU228100 |
| C. gloeosporioides | CBS 112999 | JX010152 | JX009531 | JX009818 | JX010056 | JX010445 |
| C. theobromicola | ICMP 18649 | JX010294 | JX009444 | JX009869 | JX010006 | JX010447 |
| C. truncatum | CBS 151.35 | GU227862 | GU227960 | GU228352 | GU228254 | GU228156 |
| Monilochaetes infuscans | CBS 869.96 | JQ005780 | JQ005843 | JQ005801 | JX546612 | JQ005864 |

Fig. 5 Microstructure characteristics of the pathogen of Allium ascalonicum anthracnose A,B:Conidiophores with conidia on the top;C:Conidia;D:Appressoria;E,F:Seta;G:Acervuli on the leaf of A. ascalonicum.
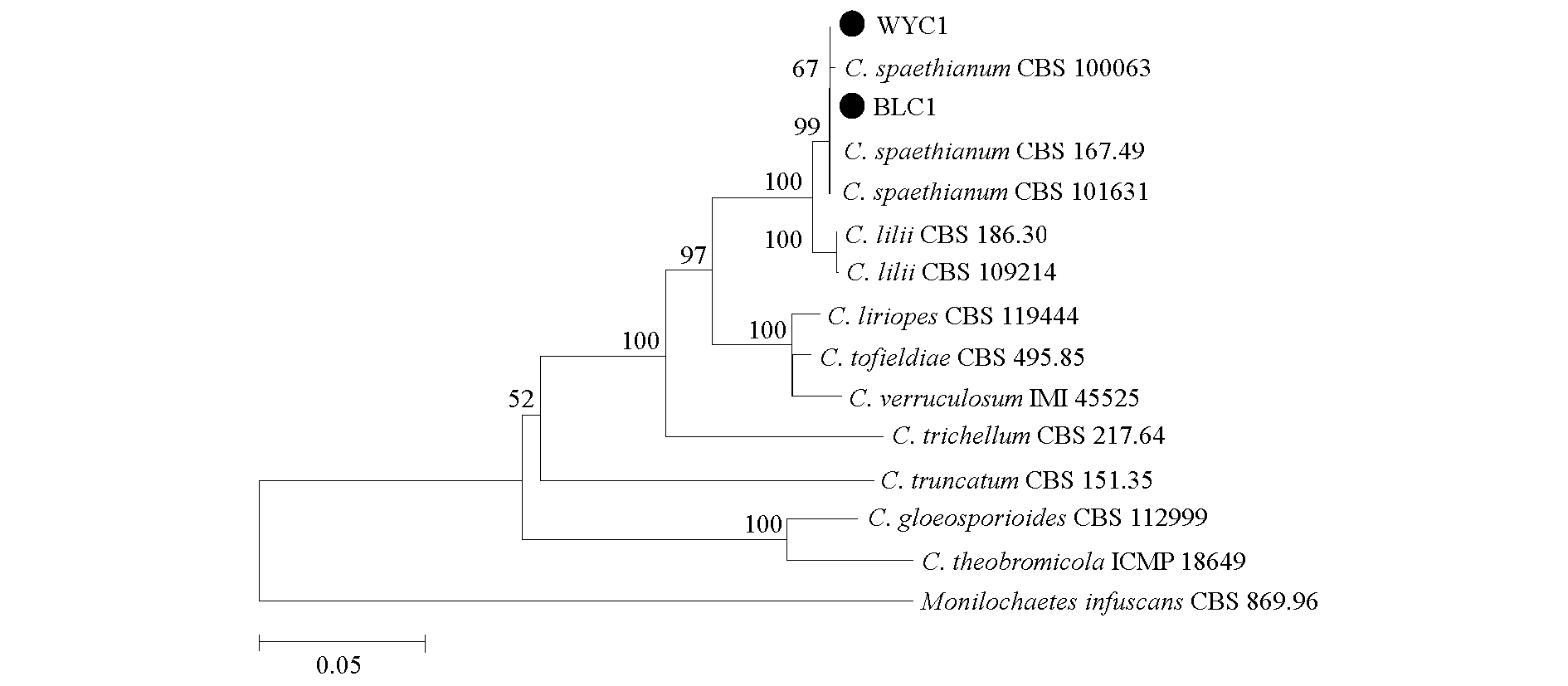
Fig. 6 Phylogenetic tree based on sequences of ITS,ACT,CHS1,GAPDH and TUB2 genes Values at the node of each clade are parsimony bootstrap above 50%,● indicates test strains.
| [1] |
Aveskamp M M, Verkley G J M, de Gruyter J, Murace M A, Perelló A, Woudenberg J H C, Groenewald J Z, Crous P W. 2009. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia, 101:363-382.
pmid: 19537209 |
| [2] | Cai L, Hyde K D, Taylor P W J, Weir B S, Waller J M, Abang M M, Zhang J Z, Yang Y L, Phoulivong S, Liu Z Y, Prihastuti H, Shivas R G, McKenzie E H C, Johnston P R. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity, 39:183-204. |
| [3] |
Cannon P F, Damm U, Johnston P R, Weir B S. 2012. Colletotrichum - current status and future directions. Studies in Mycology, 73 (1):181-213.
doi: 10.3114/sim0014 pmid: 23136460 |
| [4] |
Carbone I, Kohn L M. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia, 91 (3),553-556.
doi: 10.2307/3761358 URL |
| [5] | Chen Ai-chang, Wei Zhou-quan, Liu Xiao-juan. 2016. First report of anthracnose disease on Allium fistulosum in Gansu Province. Proceedings of the 2016 Annual Conference of the Chinese Society of Plant Pathology. Nanjing: China Agricultural Science and Technology Press: 6. (in Chinese) |
| 陈爱昌, 魏周全, 刘小娟. 2016. 大葱炭疽病在甘肃省首次发生报道. 中国植物病理学会2016 年学术年会论文集. 南京: 中国农业科学技术出版社: 6. | |
| [6] | Chen Xin-qi, Xu Jie-mei, Liang Song-yun, Tsi Zhan-he, Lang Kai-yong, Mao Zu-mei, Xu Lang-ran. 1980. Flora reipublicae popularis sinicae:Tomus 14. Angiospermae Monocotyledoneae Liliaceae (1). Beijing: Science Press: 258. (in Chinese) |
| 陈心启, 许介眉, 梁松筠, 吉占和, 郎楷永, 毛祖美, 许朗然. 1980. 中国植物志(第十四卷)被子植物门单子叶植物纲百合科(一). 北京: 科学出版社: 258. | |
| [7] | Damm U, Woudenberg J H C, Cannon P F, Crous P W. 2009. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity, 39:45-87. |
| [8] |
Diao Y, Zhang C, Liu F, Wang W, Liu L, Cai L, Liu X. 2017. Colletotrichum species causing anthracnose disease of chili in China. Persoonia, 38:20-37.
doi: 10.3767/003158517X692788 pmid: 29151625 |
| [9] | Fan Ji-de, Lu Xin-juan, Liu Can-yu, Zhao Yong-qiang, Zhang Bi-wei, Yang Hai-feng, Yang Feng. 2020. Development status and countermeasures of onion and garlic vegetable industry in Jiangsu Province. Journal of Changjiang Vegetables,(8):24-26. (in Chinese) |
| 樊继德, 陆信娟, 刘灿玉, 赵永强, 张碧薇, 杨海峰, 杨峰. 2020. 江苏省葱蒜类蔬菜产业发展现状及对策建议. 长江蔬菜,(8):24-26. | |
| [10] | Fang Zhong-da. 2007. Plant disease research methods.3rd ed. Beijing: China Agriculture Press:124-125. (in Chinese) |
| 方中达. 2007. 植病研究方法. 第3版. 北京: 中国农业出版社:124-125. | |
| [11] |
Galván G A, Wietsma W A, Putrasemedja S, Permadi A H, Kik C. 1997. Screening for resistance to anthracnose (Colletotrichum gloeosporioides Penz.) in Allium cepa and its wild relatives. Euphytica, 95:173-178.
doi: 10.1023/A:1002914225154 URL |
| [12] |
Gardes M, Bruns T D. 1993. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2:113-118.
pmid: 8180733 |
| [13] |
Ghabrial S A, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annual Review of Phytopathology, 47:353-384.
doi: 10.1146/annurev-phyto-080508-081932 pmid: 19400634 |
| [14] | Guan Y M, Liu Z B, Li M J, Wang Q X, Zhang Y Y. 2018. First report of Colletotrichum spaethianum causing anthracnose in Atractylodes japonica in China. Plant Disease, 102 (1):239. |
| [15] |
Guerber J C, Liu B, Correll J C, Johnston P R. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns,mtDNA and intron RFLPs,and mating compatibility. Mycologia, 95 (5):872-895.
pmid: 21148995 |
| [16] | Guo M, Pan Y M, Dai Y L, Gao Z M. 2013. First report of leaf spot caused by Colletotrichum spaethianum on Peucedanum praeruptorum in China. Plant Disease, 97 (10):1380. |
| [17] | Hay F, Vaghefi N, Strickland D, Hadad R, Pethybridge S. 2018. First report of Colletotrichum fioriniae causing anthracnose of elephant garlic (Allium ampeloprasum var. ampeloprasum)in New York,USA. New Disease Reports, 38:1. |
| [18] | Jia Ju-sheng, Fang De-li. 2000. Anthracnose disease of onion and its control methods. Xinjiang Agricultural Sciences,(4):171-172. (in Chinese) |
| 贾菊生, 方德立. 2000. 洋葱炭疽病及其防治措施. 新疆农业科学,(4):171-172. | |
| [19] |
Kim W G, Hong S K, Kim J H. 2008. Occurrence of Anthracnose on Welsh Onion caused by Colletotrichum circinans. Mycobiology, 36 (4):274-276.
doi: 10.4489/MYCO.2008.36.4.274 URL |
| [20] |
Liu L, Zhang L, Qiu P, Wang Y, Liu Y, Li Y, Gao J, Hsiang T. 2020. Leaf spot of Polygonatum odoratum caused by Colletotrichum spaethianum. Journal of General Plant Pathology, 86:157-161.
doi: 10.1007/s10327-019-00903-4 URL |
| [21] | Matos K S, Santana K F A, Catarino A M, Hanada R E, Silva G F. 2017. First report of anthracnose on welsh onion(Allium fistulosum)in Brazil caused by Colletotrichum theobromicola and C.truncatum. Plant Disease, 101 (6):1055. |
| [22] | Nischwitz C, Langston D, Sanders H F, Torrance R, Lewis K J, Gitaitis R D. 2008. First report of Colletotrichum gloeosporioides causing‘twister disease’of onion(Allium cepa)in Georgia. Plant Disease, 92 (6):974. |
| [23] |
O’ Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7:103-116.
doi: 10.1006/mpev.1996.0376 URL |
| [24] | Okorley B A, Sossah F L, Dan D, Li Y, Fu Y. 2019. First report of Colletotrichum spaethianum causing anthracnose on Anemarrhena asphodeloides in China. Plant Disease, 103 (6):1414. |
| [25] |
Pearson M N, Beever R E, Boine B, Arthur K. 2009. Mycoviruses of filamentous fungi and their relevance to plant pathology. Molecular Plant Pathology, 10:115-128.
doi: 10.1111/mpp.2009.10.issue-1 URL |
| [26] | Rodriguez-Salamanca L M, Enzenbacher T B, Derie M L, du Toit L J, Feng C, Correll J C, Hausbeck M K. 2012. First report of Colletotrichum coccodes causing leaf and neck anthracnose on onions(Allium cepa)in Michigan and the United States. Plant Disease, 96 (5):769. |
| [27] |
Russo V M, Pappelis A J. 1981. Observations of Colletotrichum dematium f.circinans on Allium cepa:halo formation and penetration of epidermal walls. Physiological Plant Pathology, 19 (1):127-136.
doi: 10.1016/S0048-4059(81)80014-8 URL |
| [28] | Salunkhe V N, Anandhan S, Gawande S J, Ikkar R B, Bhagat Y S, Mahajan V. 2018a. First report of Colletotrichum truncatum causing anthracnose of mouse garlic(Allium angulosum)in India. Plant Disease, 102 (1):240. |
| [29] | Salunkhe V N, Anandhan S, Gawande S J, Ikkar R B, Bhagat Y S, Mahajan V, Singh M. 2018b. First report of anthracnose caused by Colletotrichum spaethianum on Allium ledebourianum in India. Plant Disease, 102 (10):2031. |
| [30] | Santana K F A, Garcia C B, Matos K S, Hanada R E, Silva G F, Sousa N R. 2016. First report of anthracnose caused by Colletotrichum spaethianum on Allium fistulosum in Brazil. Plant Disease, 100 (1):224. |
| [31] | Sikirou R, Beed F, Hotègni J, Winter S, Assogba-Komlan F, Reeder R, Miller S A. 2011. First report of anthracnose caused by Colletotrichum gloeosporioides on onion(Allium cepa)in Benin. New Disease Reports, 23:7. |
| [32] | Sydow P. 1897. Beiträge zur Kenntnis der Pilzflora der Mark Brandenburg. I. Hedwigia Beiblatt, 36:157-164. |
| [33] | Tong Xian-ming, Wang Zheng-yi, Shi Zhi-long. 1998. Studies on the pathogen and the biological characteristics of scallion anthracnose. Acta Phytophylacica Sinica, 25 (3):249-252. (in Chinese) |
| 童贤明, 王政逸, 施志龙. 1998. 浙江省舟山市矗头炭疽病病原及其生物学特性研究. 植物保护学报, 25 (3):249-252. | |
| [34] | Vieira W A S, Michereff S J, Oliveira A C, Santos A, Câmara M P S. 2014. First report of anthracnose caused by Colletotrichum spaethianum on Hemerocallis flava in Brazil. Plant Disease, 98 (7):997. |
| [35] | White T J, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics//Innis M A,Gelfand D H,Sninsky J J,White T J. PCR Protocols:a guide to methods and applications. San Diego,California,U.S.A: Academic Press:315-322. |
| [36] | Wu Yahong, Zhao Qing, Wang Haiping. 2021. Research progress on the classification of Allium plants. Acta Horticulturae Sinica, 48 (7):1418-1428. (in Chinese) |
| 武亚红, 赵青, 王海平. 2021. 葱属植物分类研究进展. 园艺学报, 47 (7):1418-1428. | |
| [37] | Yang L, Lu X H, Jing Y L, Li S D, Wu B M. 2019. First report of common bean(Phaseolus vulgaris)stem rot caused by Colletotrichum spaethianum in China. Plant Disease, 103 (1):151. |
| [38] |
Yang Y, Liu Z, Cai L, Hyde K D. 2012. New species and notes of Colletotrichum on daylilies(Hemerocallis spp.). Tropical Plant Pathology, 37 (3):165-174.
doi: 10.1590/S1982-56762012000300001 URL |
| [39] |
Yu L, Sang W, Wu M, Zhang J, Yang L, Zhou Y, Chen W, Li G. 2015. Novel hypovirulence-associated RNAmycovirus in the plant-pathogenic fungus Botrytis cinerea:molecular and biological characterization. Applied and Environment Microbiology, 81 (7):2299-2310.
doi: 10.1128/AEM.03992-14 URL |
| [40] | Yu Lin, She Xiao-man, Lan Guo-bing, Tang Ya-fei, Li Zheng-gang, Deng Ming-guang, He Zi-fu. 2020. Identification of pathogen causing sweet potato scab in Guangdong. Journal of Southern Agriculture, 51 (3):579-585. (in Chinese) |
| 于琳, 佘小漫, 蓝国兵, 汤亚飞, 李正刚, 邓铭光, 何自福. 2020. 广东甘薯疮痂病病原菌鉴定. 南方农业学报, 51 (3):579-585. | |
| [41] | Zeng Da-xing, Qi Pei-kun, Jiang Zi-de. 2002. Identification of the falcate Colletotrichum on vegetable using RAPD analysis. Acta Phytopathologica Sinica, 32 (2):170-174. (in Chinese) |
| 曾大兴, 戚佩坤, 姜子德. 2002. 应用RAPD技术快速鉴定蔬菜上的弯孢类炭疽菌. 植物病理学报, 32 (2):170-174. | |
| [42] | Zhao W, Wang T, Chen Q Q, Chi Y K, Swe T M, Qi R D. 2016. First report of leaf spot caused by Colletotrichum spaethianum on Lilium lancifolium in China. Plant disease, 100 (11):2328. |
| [1] | LIANG Jiali, WU Qisong, CHEN Guangquan, ZHANG Rong, XU Chunxiang, and FENG Shujie, . Identification of the Neopestalotiopsis musae Pathogen of Banana Leaf Spot Disease [J]. Acta Horticulturae Sinica, 2023, 50(2): 410-420. |
| [2] | GONG Changyi, LIU Jiaojiao, DENG Qiang, ZHANG Lixin. Identification and Pathogenicity of Colletotrichum Species Causing Anthracnose on Camellia sinensis [J]. Acta Horticulturae Sinica, 2022, 49(5): 1092-1101. |
| [3] | ZHANG Xiaoyong, LI Shujiang, YAN Kai, WENG Guiying, ZHANG Yunxia, LONG Qiaofang, LIANG Wen, JIANG Qinna, YANG Youlian. Screening and Culture Characteristics of Biocontrol Strains of Postharvest Anthracnose of Mango [J]. Acta Horticulturae Sinica, 2021, 48(11): 2171-2184. |
| [4] | WANG Tongtong,GAO Fen,LIU Dan,and LI Peili*. Primary Identification of the Pathogen Causing Anthracnose of Nandina domestica in Chengdu,Sichuan [J]. ACTA HORTICULTURAE SINICA, 2019, 46(8): 1576-1584. |
| [5] | LI Jiawei,XU Lanyi,WANG Dongshuang,Lü Lijie,CHEN Xiaomeng,and ZHANG Dongdong*. Screening and Identification of Bacillus Antagonist Against Anthracnose of Jujube and Analysis of Its Antifungal Substances [J]. ACTA HORTICULTURAE SINICA, 2019, 46(12): 2406-2414. |
| [6] | XU Guoliang1,Zhao Feifei1,WANG Qinglian2,WANG Shiling1,JIA Jin1,CHEN Yanling2,HUANG Chuanhong3,LIU Yongzhong2,PENG Shu’ang2,WANG Guoping1,HONG Ni1,and DING Fang1,2,*. Identification of a New Pathogen of Citrus Postharvest Disease [J]. ACTA HORTICULTURAE SINICA, 2018, 45(10): 2037-2044. |
| [7] | CHENG Lin1,*,ZHANG Sheng1,*,LI Yan-qing2,CHEN Fu-dong1,CHENG Fei3,ZHANG Xiao-yan1,DONG Tian1,and GUO Jia-jin1,**. Pathogen Identification of Fusarium Crown Root Rot and Screening for Resistant Sources in Tomato [J]. ACTA HORTICULTURAE SINICA, 2016, 43(4): 781-788. |
| [8] | LI Li1,SONG Shu-xiang1,LIU Hui-xiang2,and GUO Xian-feng1,*. Identification of Red Spot Pathogens on Peony in Shandong Province [J]. ACTA HORTICULTURAE SINICA, 2016, 43(2): 365-372. |
| [9] | GAO Xiao-ming1,2,LIU Chong-huai1,ZHANG Guo-hai2,ZHANG Ying1,JIANG Jian-fu1,SUN Hai-sheng1,and FAN Xiu-cai1,*. Preliminary Analysis of Grape Anthracnose Resistance QTL [J]. ACTA HORTICULTURAE SINICA, 2016, 43(12): 2442-2450. |
| [10] | ZHOU Ping-lan1,4,TANG Xin-ke4,LONG Yue-lin2,*,and ZHOU Xiao-mao2,3,*. Identification of Pathogen Colletotrichum boninense from Cymbidium goeringii and Its Biological Characteristics [J]. ACTA HORTICULTURAE SINICA, 2015, 42(10): 1993-2001. |
| [11] | ZHAO Xi-ting1,2,3,*,WANG Miao1,WANG Tian-le1,and LI Ming-jun1,2,3. Isolation and Identification of Pathogen Causing Black Spot Disease of Chrysanthemum morifolium‘Huaihuang’ [J]. ACTA HORTICULTURAE SINICA, 2015, 42(1): 174-182. |
| [12] | SUN Chun-Ying, MAO Sheng-Li, ZHANG Zheng-Hai, WANG Li-Hao, ZHANG Bao-Xi-*. Progress on Genetics and Breeding of Resistance to Anthracnose (Colletotrichum spp.)in Pepper [J]. ACTA HORTICULTURAE SINICA, 2013, 40(3): 579-590. |
| [13] | GONG De-Qiang, GU Hui, ZHANG Lu-Bin, HONG Ke-Qian, ZHU Shi-Jiang. Effects of Pretharvest Methyl Jasmonate Spraying on Disease Resistance and Postharvest Quality of Mango Fruits [J]. ACTA HORTICULTURAE SINICA, 2013, 40(1): 49-57. |
| [14] | YANG Xiu-mei;QU Su-ping;WANG Li-hua;LIU Xiao-xia;PENG Lü-chun;and WANG Ji-hua;. Identification and PCR Detection of Phytophthora Blight of Lily [J]. ACTA HORTICULTURAE SINICA, 2011, 38(6): 1180-1184. |
| [15] | GU Yu;HAN Qi-hou;WANG Wu-tai;LI Su-wen;SUN De-ling;and WU Feng . Application of Common Bean Anthracnose Resistance Gene SCAR Markers in Snap Bean Disease Resistance Identification [J]. ACTA HORTICULTURAE SINICA, 2011, 38(5): 911-920. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd