
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (12): 3271-3287.doi: 10.16420/j.issn.0513-353x.2024-0990
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
HAN Hongwei, CHANG Yanan, LIU Huifang, ZHUANG Hongmei, XING Jiayi, ZHANG Xiaoda, ZULIPIYE · Abudumilike, WANG Hao*( ), WANG Qiang*
), WANG Qiang*
Received:2025-01-05
Revised:2025-09-03
Online:2025-12-25
Published:2025-12-20
Contact:
WANG Hao, WANG Qiang
HAN Hongwei, CHANG Yanan, LIU Huifang, ZHUANG Hongmei, XING Jiayi, ZHANG Xiaoda, ZULIPIYE · Abudumilike, WANG Hao, WANG Qiang. Identification of Cytochrome P450 Family Genes in Melon and Expression Analysis in Response to Light Intensity[J]. Acta Horticulturae Sinica, 2025, 52(12): 3271-3287.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0990
| 处理 Treatment | 方法 Method | 平均透光率/% Average transmittance | 光辐射强度(× 104 lx) Light radiation intensity |
|---|---|---|---|
| 强光(SW) Strong light | 自然光(无遮光) Natural light(without shading) | 100.0 | 6.08 |
| 亚强光(SN) Sub-strong light | 距离植株生长点上方0.30 m覆盖一层棚膜 A layer of greenhouse film was covered 0.30 m above the plant growth point | 78.2 | 4.75 |
| 遮荫(ZY) Shading | 在SN处理的基础上再覆盖一层遮阳网 Based on the SN treatment,an additional layer of shading net was covered | 28.7 | 1.74 |
Table 1 Transmittance and light radiation intensity of different treatments
| 处理 Treatment | 方法 Method | 平均透光率/% Average transmittance | 光辐射强度(× 104 lx) Light radiation intensity |
|---|---|---|---|
| 强光(SW) Strong light | 自然光(无遮光) Natural light(without shading) | 100.0 | 6.08 |
| 亚强光(SN) Sub-strong light | 距离植株生长点上方0.30 m覆盖一层棚膜 A layer of greenhouse film was covered 0.30 m above the plant growth point | 78.2 | 4.75 |
| 遮荫(ZY) Shading | 在SN处理的基础上再覆盖一层遮阳网 Based on the SN treatment,an additional layer of shading net was covered | 28.7 | 1.74 |
| 引物名称Primer name | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|
| MELO3C021434.2 | GGTTCAGGCTTACAAGGCTAAT | CCACTCTCAAGGCATTCTACG |
| MELO3C009884.2 | GGTGCCACTATTGTGTCTCTTC | GCCTATTCGGTTAGCAACTCTT |
| MELO3C002338.2 | AAGCTCTTCCAATGCACCAAG | TGATGCCACAACACCAACAC |
| MELO3C018724.2 | AGCATGGCAGGATTAGGAATTG | TTGTGGCGTTGGAGGAGTT |
| MELO3C011682.2 | CATCTCGGCGTGGAAGGTAT | CGTGGCTGTCGTTCAAGTG |
| MELO3C011976.2 | CGGCGGAGTATTGTCAGATG | CAACCAATCCAAGAACAACACA |
| MELO3C007154.2 | TGAGCCACCATCCTACCTAAC | CACTTCCAACTCACGGTTCTT |
| MELO3C004214.2 | GGAGAAGTGCCAGAGAACCT | TTTCACCCAGTTGCCCAATC |
| MELO3C022113.2 | CGAGGCTGTCTGGTTCAAGA | CAATGACGGCGACGATGAG |
| MELO3C027057.2 | GCAGACGCCAACGCCATTA | GCACGATCCAATTACGACGATT |
| MELO3C004135.2 | CGGCAGCAACATTGAAACAA | TCTCACAAGCAGCAACAACA |
| MELO3C004201.2 | ACACGACTTGGACTTGGAGAG | CCACAATGCTCAACAGAGACAA |
| MELO3C002187.2 | CAATAGTCAGTGGCAACAACCT | GCGTAGCAGTTTCCGAATCAT |
| MELO3C006499.2 | GGACAACGACTAATCGCCTAC | ATGATGGAATCGCTTGCTCTC |
| MELO3C026749.2 | GGATGTGGTAGACTCTGTAGCA | CTTGACTGTCCTCCTGAGATTC |
| MELO3C026019.2 | TTCCATTAACCAGTGCTGCTC | GCTCTTGACGAAGTGCTTGTA |
| MELO3C016167.2 | TGTTGCCGTCCGATCCTTAT | CTTCCTCTTGCTCCTCTCCTT |
| MELO3C035538.2(CmCYP195) | TTGTCCAACTTGTTAGCGATGT | CTTGAATCCCGTGAAGATGGTA |
| MELO3C004801.2(CmCYP24) | GTCCAATCCTTCGCCTTCATT | GCCATTCGACAGATCACATCAT |
| MELO3C026260.2(CmCYP174) | TGCTTCCTCTTCTTCTTCTTCC | TTGGTGGAGATGGAGGTGAA |
| MELO3C011869.2(CmCYP79) | CACCGCTTCTCTGTCAACCT | TGAGCAATGGCAGCAAGATTC |
| MELO3C021842.2(CmCYP144) | ATCACTTCCAACCAACACAACA | CTGTAGGACCTGACACGAGAA |
| MELO3C015058.2(CmCYP101) | CCAGCACCACTGAGATATTGAG | TGAGAACATGAGAAGCAAGGAT |
| MELO3C013868.2(CmCYP95) | GCTTCGACGGCTGATAACAAT | GCAACTCCAATGGCTTCTCC |
Table 2 Pirmer sequences used for qRT-PCR analysis
| 引物名称Primer name | 上游引物(5′-3′)Forward primer | 下游引物(5′-3′)Reverse primer |
|---|---|---|
| MELO3C021434.2 | GGTTCAGGCTTACAAGGCTAAT | CCACTCTCAAGGCATTCTACG |
| MELO3C009884.2 | GGTGCCACTATTGTGTCTCTTC | GCCTATTCGGTTAGCAACTCTT |
| MELO3C002338.2 | AAGCTCTTCCAATGCACCAAG | TGATGCCACAACACCAACAC |
| MELO3C018724.2 | AGCATGGCAGGATTAGGAATTG | TTGTGGCGTTGGAGGAGTT |
| MELO3C011682.2 | CATCTCGGCGTGGAAGGTAT | CGTGGCTGTCGTTCAAGTG |
| MELO3C011976.2 | CGGCGGAGTATTGTCAGATG | CAACCAATCCAAGAACAACACA |
| MELO3C007154.2 | TGAGCCACCATCCTACCTAAC | CACTTCCAACTCACGGTTCTT |
| MELO3C004214.2 | GGAGAAGTGCCAGAGAACCT | TTTCACCCAGTTGCCCAATC |
| MELO3C022113.2 | CGAGGCTGTCTGGTTCAAGA | CAATGACGGCGACGATGAG |
| MELO3C027057.2 | GCAGACGCCAACGCCATTA | GCACGATCCAATTACGACGATT |
| MELO3C004135.2 | CGGCAGCAACATTGAAACAA | TCTCACAAGCAGCAACAACA |
| MELO3C004201.2 | ACACGACTTGGACTTGGAGAG | CCACAATGCTCAACAGAGACAA |
| MELO3C002187.2 | CAATAGTCAGTGGCAACAACCT | GCGTAGCAGTTTCCGAATCAT |
| MELO3C006499.2 | GGACAACGACTAATCGCCTAC | ATGATGGAATCGCTTGCTCTC |
| MELO3C026749.2 | GGATGTGGTAGACTCTGTAGCA | CTTGACTGTCCTCCTGAGATTC |
| MELO3C026019.2 | TTCCATTAACCAGTGCTGCTC | GCTCTTGACGAAGTGCTTGTA |
| MELO3C016167.2 | TGTTGCCGTCCGATCCTTAT | CTTCCTCTTGCTCCTCTCCTT |
| MELO3C035538.2(CmCYP195) | TTGTCCAACTTGTTAGCGATGT | CTTGAATCCCGTGAAGATGGTA |
| MELO3C004801.2(CmCYP24) | GTCCAATCCTTCGCCTTCATT | GCCATTCGACAGATCACATCAT |
| MELO3C026260.2(CmCYP174) | TGCTTCCTCTTCTTCTTCTTCC | TTGGTGGAGATGGAGGTGAA |
| MELO3C011869.2(CmCYP79) | CACCGCTTCTCTGTCAACCT | TGAGCAATGGCAGCAAGATTC |
| MELO3C021842.2(CmCYP144) | ATCACTTCCAACCAACACAACA | CTGTAGGACCTGACACGAGAA |
| MELO3C015058.2(CmCYP101) | CCAGCACCACTGAGATATTGAG | TGAGAACATGAGAAGCAAGGAT |
| MELO3C013868.2(CmCYP95) | GCTTCGACGGCTGATAACAAT | GCAACTCCAATGGCTTCTCC |

Fig. 6 Gene replication and collinearity analysis of CmCYP450 The collinearity relationships of gene pairs are represented by colorful lines. Different chromosomes are represented by different colors

Fig. 7 Synteny analysis of P450 genes between the genomes of Cucumis melo,Arabidopsis thaliana and Oryza sativa The gray lines are all collinear relationships among different genomes,and the colored lines are collinear relationships among P450 gene family genes
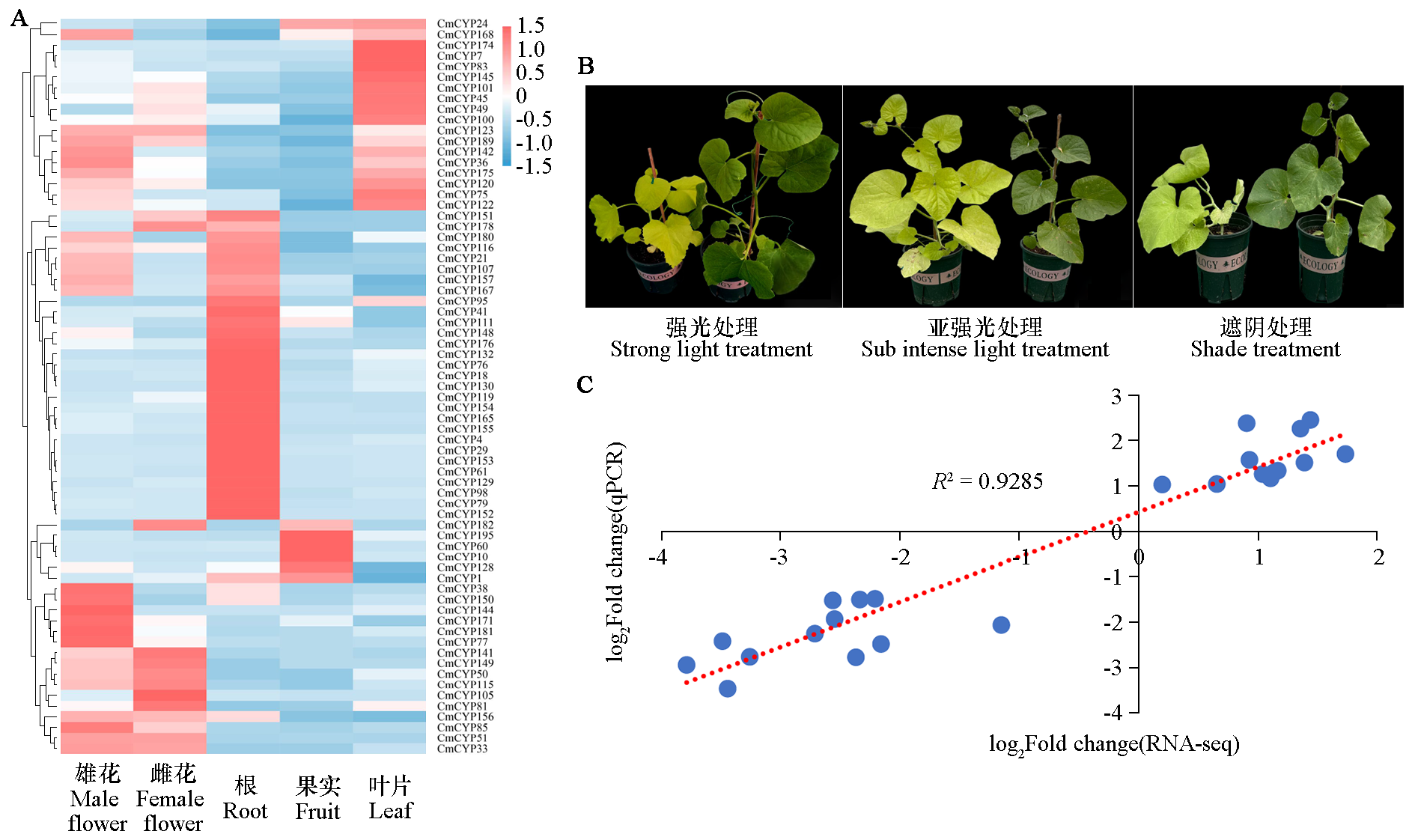
Fig. 9 The heat map of differential expression of CmCYP450 under different light radiation intensities(A),Phenotypes of mutants and wild-type plants(B),Correlation analysis of relative expression levels between RNA seq and qRT PCR(C) The plant on the left in Figure B is the mutant Cmygl-1,and the plant on the right is the wild type
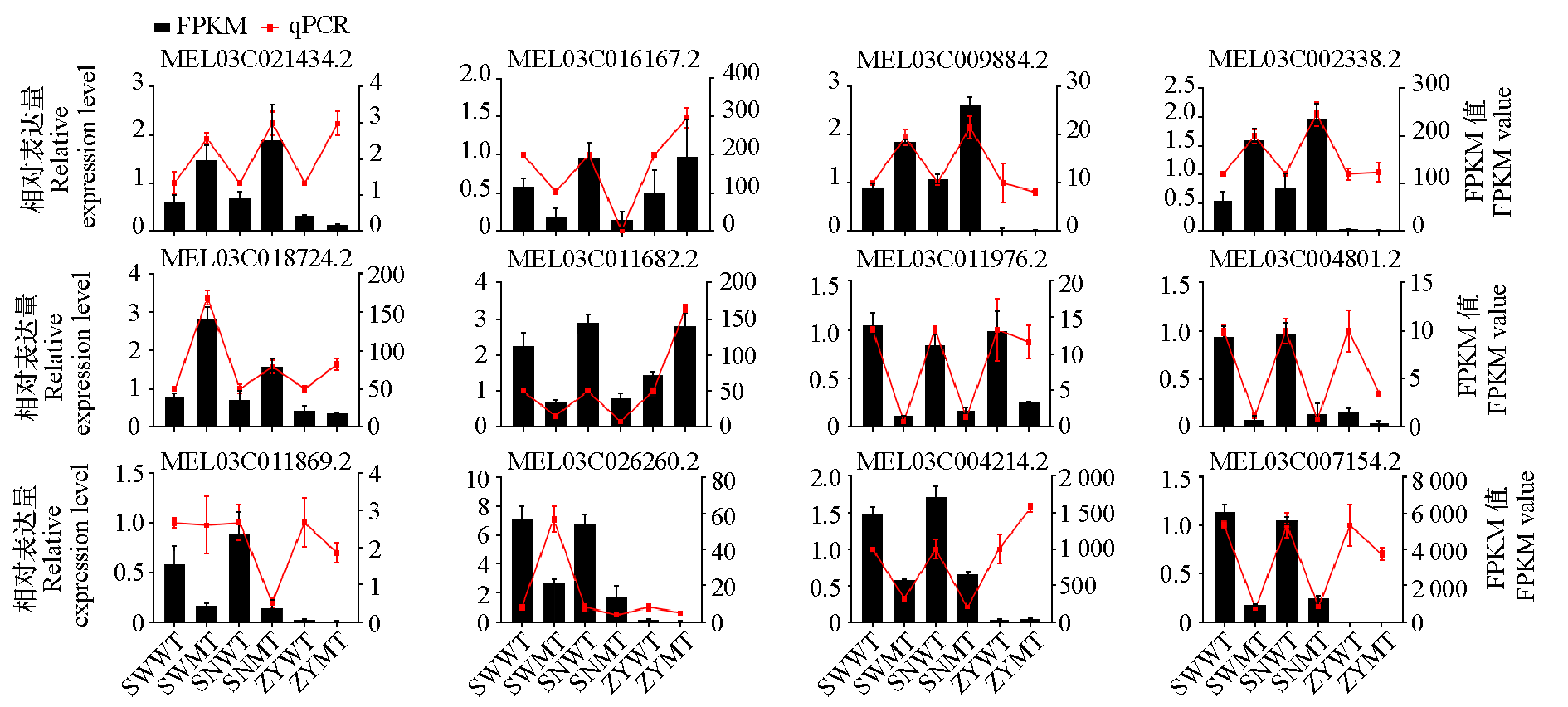
Fig. 10 Analysis of expression trends of some CmCYP450 genes using qRT-PCR(fold change)and RNA-seq(FPKM) SWWT:The wild type plants with strong light treatment;SNWT:The wild type plants with sub intense light treatment;ZYWT:The wild type plants with shade treatment;SNMT:The mutants with strong light treatment;SNMT:The mutants with sub intense light treatment;ZYMT:The mutants with shade treatment
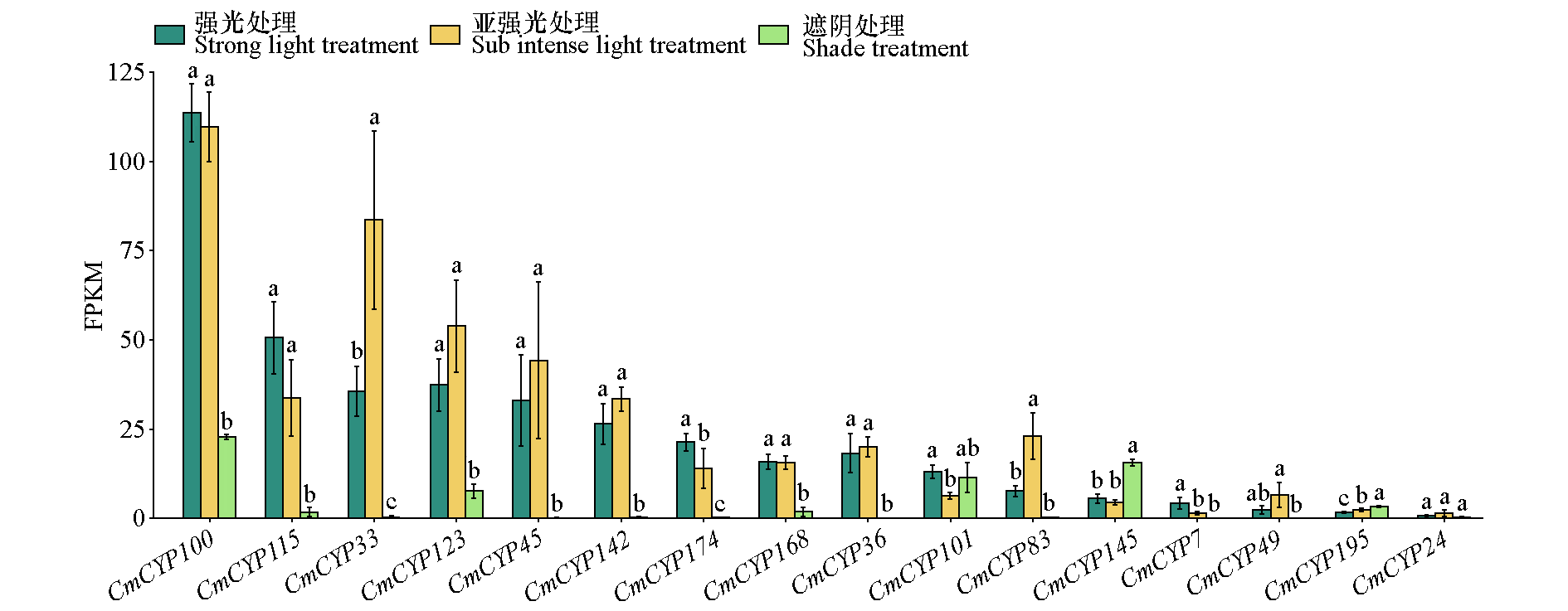
Fig. 11 The expression trends of 16 CmCYP450 genes highly expressed in mutant leaves in response to different light radiation intensities FPKM:Fragments Per Kilobase of transcript per Million mapped reads. Different lowercase letter indicate significant differences at 0.05 level
| [1] |
|
| [2] |
|
|
曹冠华, 张兴开, 柏旭, 毕悦, 殷红伟, 张芸, 贺森. 2022. 三七细胞色素P450酶基因PnCyp450_3响应丛枝菌根真菌诱导的表达特性分析. 中草药, 53(21):6848-6856.
|
|
| [3] |
|
| [4] |
|
| [5] |
|
|
崔会婷. 2018. 蒺藜苜蓿细胞色素P450基因的克隆及功能分析[硕士论文]. 北京: 中国农业科学院.
|
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
|
李万青, 高波, 杨俊, 陈鹏, 李玉红. 2015. 一个新的黄瓜叶色黄化突变体的生理特性分析. 西北农业学报, 24(7):98-103.
|
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [1] | WANG Baochen, LUO Chen, YAN Jinqiang, CHENG Xiaoxin, MO Renlian, LIU Wenrui, ZHANG Yuyang, JIANG Biao. Cloning,Expression,and Functional Research of BhYAB4b in Wax Gourd [J]. Acta Horticulturae Sinica, 2025, 52(9): 2353-2362. |
| [2] | ZHANG Ting, WANG Botao, ZHANG Lei, SONG Lihua, CAO Bing, MA Yaping. Identification of the LbWaxy Gene Family in Lycium barbarum and Analysis of Gene Expression Characteristics Under High Concentrations of CO2 [J]. Acta Horticulturae Sinica, 2025, 52(9): 2395-2409. |
| [3] | ZHANG Tuo, NIE Menghan, LIU Yuanyuan, DUAN Xinlian, HAN Xing, XIE Baogui, and YAN Junjie. Expression Analysis of the Catalase Genes FfCAT1 and FfCAT2 in Flammulina filiformis [J]. Acta Horticulturae Sinica, 2025, 52(7): 1769-1779. |
| [4] | SUN Tingzhen, SHU Qin, MA Wei, SHI Yuzi, ZHANG Meng, XIANG Chenggang, BO Kailiang, DUAN Ying, WANG Changlin. Identification and Phylogenetic Analysis of Gibberellin Oxidase Gene Family and Its Expression Pattern in Main Vine Growth in Cucurbita maxima [J]. Acta Horticulturae Sinica, 2025, 52(3): 603-622. |
| [5] | DENG Shuqin, GAO Yingrui, LI Yutong, WANG Ying, GONG Chunmei, BAI Juan. Response of Ubiquitin-ligase Gene CsPUB21 to Different Abiotic Stress in Camellia sinensis [J]. Acta Horticulturae Sinica, 2025, 52(3): 655-670. |
| [6] | TONG Zongjun, HAN Xing, DUAN Xinlian, LIU Yuanyuan, LIN Junbin, GAN Ying, CHEN Jie, XIE Baogui, GAN Bingcheng, YAN Junjie. Sequence Characterization of PRX Family and Expression Analysis in the Process of Fruiting Body Development in Flammulina filiformis [J]. Acta Horticulturae Sinica, 2025, 52(2): 337-348. |
| [7] | LIU Lu, WANG Meng, ZHENG Xiaomei, WANG Keqing, MIAO Ruyi, ZANG Qiaolu. Identification and Expression Analysis of CBF Gene Family of Artemisia argyi [J]. Acta Horticulturae Sinica, 2025, 52(10): 2703-2712. |
| [8] | HE Dandan, HE Hongtai, WANG Wenting, ZHOU Wenmei, LIU Yanmin, LIU Sushuang. Identification of Melon GolS Genes Family and the Expression Analysis in Response to Low Temperature Stress [J]. Acta Horticulturae Sinica, 2025, 52(1): 136-148. |
| [9] | GUO Changquan, LI Danqi, HUI Xinran, ZHENG Jingya, HOU Menglu, ZHU Yongxing. Identification and Expression Pattern Analysis of DUF966 Gene Family Members in Ginger [J]. Acta Horticulturae Sinica, 2024, 51(9): 2031-2047. |
| [10] | ZHAO Jiaying, ZENG Zhouting, CEN Xinying, SHI Jiaoqi, LI Xiaoxian, SHEN Xiaoxia, YU Zhenming. Genome-Wide Identification and Expression Analysis of CCO Gene Family in Dendrobium officinale During Flower Development [J]. Acta Horticulturae Sinica, 2024, 51(9): 2075-2088. |
| [11] | LIU Yanyan, DING Ying, LIU Xinghua, ZHENG Jiaqiu, LIU Zhiqin. Identification of Pepper CaSYT1 and Its Function in Phytophthora capsici Infection [J]. Acta Horticulturae Sinica, 2024, 51(3): 533-544. |
| [12] | LÜ Yunzhou, DONG Xiaoyun, SUN Hainan, LIANG Zhenhai, and HUANG Libin. A New Koelreuteria paniculata Cultivar‘Caihong’ [J]. Acta Horticulturae Sinica, 2023, 50(S1): 187-188. |
| [13] | LI Yuteng, CHEN Yao, REN Hengze, LI Congcong, WANG Haoqian, CAO Hongli, YUE Chuan, HAO Xinyuan, WANG Xinchao. Identification,Expression Analysis and Interaction Validation of CsIDM in Tea Plants [J]. Acta Horticulturae Sinica, 2023, 50(8): 1679-1696. |
| [14] | CAO Xiongjun, HAN Jiayu, CHENG Guo, WANG Bo, MA Guangren, LIN Ling, TAN Zongkun, HUANG Qiumi, CHEN Xiao, CHEN Fuyi, SHI Xiaofang, PAN Fengping, BAI Xianjin. Effects of Light Duration and Intensity on Yield Formation of‘Shine- Muscat’Grape [J]. Acta Horticulturae Sinica, 2023, 50(8): 1739-1746. |
| [15] | XUE Zhenzhen, GUAN Hongfa, LI Na, LI Lingfei, ZHONG Chunmei. Genome-wide Identification of GASA Family and Preliminary Exploration of Leaf Variegation Formation in Begonia masoniana [J]. Acta Horticulturae Sinica, 2023, 50(7): 1482-1494. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd