
Acta Horticulturae Sinica ›› 2025, Vol. 52 ›› Issue (12): 3288-3302.doi: 10.16420/j.issn.0513-353x.2024-0781
• Genetic & Breeding·Germplasm Resources·Molecular Biology • Previous Articles Next Articles
PENG Chaofeng1,2, QI Shuaizheng2, GENG Xining2, YAO Pengqiang2, XIE Lihua2, ZHANG Yu2,3, CHEN Minghui2, SHI Jiang1,*( ), CHENG Shiping2,*(
), CHENG Shiping2,*( )
)
Received:2025-05-29
Revised:2025-09-26
Online:2025-12-25
Published:2025-12-20
Contact:
SHI Jiang, CHENG Shiping
PENG Chaofeng, QI Shuaizheng, GENG Xining, YAO Pengqiang, XIE Lihua, ZHANG Yu, CHEN Minghui, SHI Jiang, CHENG Shiping. Differential Expressed Analysis by Transcriptome Sequencing in Leaves of Different Ploidy Paeonia ostii‘Fengdan’[J]. Acta Horticulturae Sinica, 2025, 52(12): 3288-3302.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2024-0781
| 倍性 Ploidy | 叶长/cm Leaf length | 叶宽/cm Leaf width | 叶面积/cm2 Leaf area | 叶片厚度/μm Leaf thickness | 叶脉直径/μm Vein diameter |
|---|---|---|---|---|---|
| 二倍体 Diploid | 9.56 ± 0.50 a | 2.93 ± 0.25 b | 23.38 ± 2.08 b | 152.70 ± 15.34 c | 828.08 ± 46.42 b |
| 三倍体 Triploid | 9.03 ± 0.85 a | 5.80 ± 0.87 a | 34.86 ± 5.76 a | 206.76 ± 14.19 b | 840.70 ± 34.38 b |
| 四倍体 Tetraploid | 7.23 ± 0.25 b | 3.80 ± 0.50 b | 25.01 ± 3.01 b | 233.81 ± 18.95 a | 976.92 ± 56.21 a |
Table 1 Growth characteristics of different ploidy Paeonia ostii‘Fengdan’
| 倍性 Ploidy | 叶长/cm Leaf length | 叶宽/cm Leaf width | 叶面积/cm2 Leaf area | 叶片厚度/μm Leaf thickness | 叶脉直径/μm Vein diameter |
|---|---|---|---|---|---|
| 二倍体 Diploid | 9.56 ± 0.50 a | 2.93 ± 0.25 b | 23.38 ± 2.08 b | 152.70 ± 15.34 c | 828.08 ± 46.42 b |
| 三倍体 Triploid | 9.03 ± 0.85 a | 5.80 ± 0.87 a | 34.86 ± 5.76 a | 206.76 ± 14.19 b | 840.70 ± 34.38 b |
| 四倍体 Tetraploid | 7.23 ± 0.25 b | 3.80 ± 0.50 b | 25.01 ± 3.01 b | 233.81 ± 18.95 a | 976.92 ± 56.21 a |
| 样品 倍性 Sample Ploidy | 原始数据 Raw reads | 质控数据 Clean reads | 质控数据碱基数 Clean base(G) | Q20/% | Q30/% | GC含量/% GC Content | |
|---|---|---|---|---|---|---|---|
| 盛花期 Full flowering period | 二倍体 Diploid | 59 143 608 | 55 527 696 | 8.33 | 98.96 | 96.66 | 45.23 |
| 53 698 588 | 50 179 058 | 7.53 | 98.82 | 96.25 | 45.10 | ||
| 54 181 490 | 50 801 050 | 7.62 | 98.77 | 96.10 | 45.07 | ||
| 三倍体 Triploid | 43 391 280 | 41 104 402 | 6.17 | 98.85 | 96.31 | 45.15 | |
| 53 750 618 | 50 548 380 | 7.58 | 99.01 | 96.79 | 45.17 | ||
| 50 647 764 | 47 605 318 | 7.14 | 98.98 | 96.73 | 45.17 | ||
| 四倍体 Tetraploid | 57 476 282 | 54 146 594 | 8.12 | 98.96 | 96.66 | 45.05 | |
| 43 140 704 | 40 339 548 | 6.05 | 98.92 | 96.54 | 44.98 | ||
| 49 974 664 | 47 103 112 | 7.07 | 98.84 | 96.29 | 44.90 | ||
| 花衰 败期 Flower decay period | 二倍体 Diploid | 57 079 036 | 53 328 478 | 8.00 | 99.01 | 96.81 | 44.77 |
| 52 656 282 | 49 232 680 | 7.38 | 98.83 | 96.22 | 44.75 | ||
| 52 691 956 | 49 349 472 | 7.40 | 98.79 | 96.10 | 44.82 | ||
| 三倍体 Triploid | 58 114 202 | 54 795 360 | 8.22 | 98.91 | 96.47 | 44.95 | |
| 48 957 976 | 45 976 076 | 6.90 | 98.90 | 96.43 | 44.95 | ||
| 53 091 772 | 50 658 874 | 7.60 | 98.86 | 96.32 | 44.90 | ||
| 四倍体 Tetraploid | 57 574 644 | 55 121 600 | 8.27 | 98.76 | 96.04 | 44.88 | |
| 55 111 304 | 52 274 248 | 7.84 | 99.00 | 96.78 | 45.01 | ||
| 54 178 576 | 50 654 914 | 7.60 | 99.00 | 96.79 | 45.03 | ||
Table 2 Quality analysis of transcriptome sequencing data
| 样品 倍性 Sample Ploidy | 原始数据 Raw reads | 质控数据 Clean reads | 质控数据碱基数 Clean base(G) | Q20/% | Q30/% | GC含量/% GC Content | |
|---|---|---|---|---|---|---|---|
| 盛花期 Full flowering period | 二倍体 Diploid | 59 143 608 | 55 527 696 | 8.33 | 98.96 | 96.66 | 45.23 |
| 53 698 588 | 50 179 058 | 7.53 | 98.82 | 96.25 | 45.10 | ||
| 54 181 490 | 50 801 050 | 7.62 | 98.77 | 96.10 | 45.07 | ||
| 三倍体 Triploid | 43 391 280 | 41 104 402 | 6.17 | 98.85 | 96.31 | 45.15 | |
| 53 750 618 | 50 548 380 | 7.58 | 99.01 | 96.79 | 45.17 | ||
| 50 647 764 | 47 605 318 | 7.14 | 98.98 | 96.73 | 45.17 | ||
| 四倍体 Tetraploid | 57 476 282 | 54 146 594 | 8.12 | 98.96 | 96.66 | 45.05 | |
| 43 140 704 | 40 339 548 | 6.05 | 98.92 | 96.54 | 44.98 | ||
| 49 974 664 | 47 103 112 | 7.07 | 98.84 | 96.29 | 44.90 | ||
| 花衰 败期 Flower decay period | 二倍体 Diploid | 57 079 036 | 53 328 478 | 8.00 | 99.01 | 96.81 | 44.77 |
| 52 656 282 | 49 232 680 | 7.38 | 98.83 | 96.22 | 44.75 | ||
| 52 691 956 | 49 349 472 | 7.40 | 98.79 | 96.10 | 44.82 | ||
| 三倍体 Triploid | 58 114 202 | 54 795 360 | 8.22 | 98.91 | 96.47 | 44.95 | |
| 48 957 976 | 45 976 076 | 6.90 | 98.90 | 96.43 | 44.95 | ||
| 53 091 772 | 50 658 874 | 7.60 | 98.86 | 96.32 | 44.90 | ||
| 四倍体 Tetraploid | 57 574 644 | 55 121 600 | 8.27 | 98.76 | 96.04 | 44.88 | |
| 55 111 304 | 52 274 248 | 7.84 | 99.00 | 96.78 | 45.01 | ||
| 54 178 576 | 50 654 914 | 7.60 | 99.00 | 96.79 | 45.03 | ||
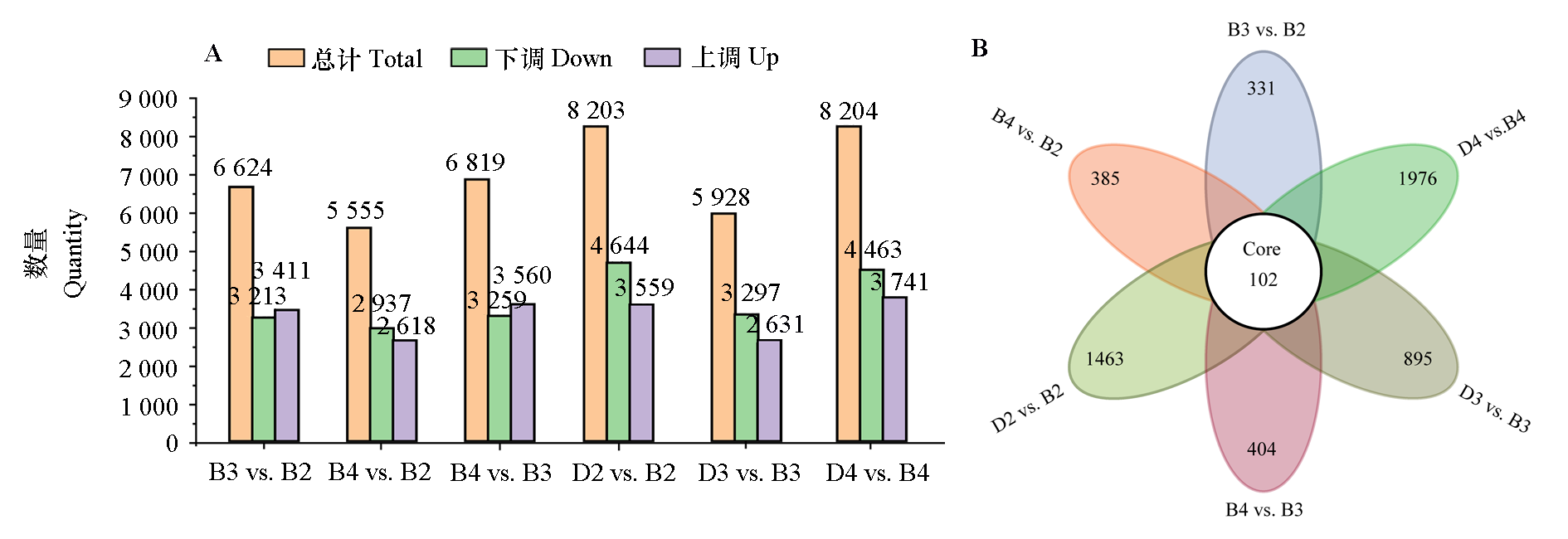
Fig. 3 Differential expression genes(DEG)(A)and venn diagram(B)in the leaves Paeonia ostii‘Fengdan’under different ploidys and periods B2:Diploid during the blooming period;B3:Triploid during the blooming period;B4:Tetraploid during the blooming period;D2:Diploid during the decline period;D3:Triploid during the decline period;D4:Tetraploid during the decline period

Fig. 4 Heatmap of the DEG in auxin signaling transduction(A),cytokinin signaling transduction(B),zeatin biosynthesis(C)and chlorophyll metabolic pathways(D)during polyploid leaf of Paeonia ostii‘Fengdan’ B2:Diploid during the blooming period;B3:Triploid during the blooming period;B4:Tetraploid during the blooming period;D2:Diploid during the decline period;D3:Triploid during the decline period;D4:Tetraploid during the decline period. The same below
| [1] |
|
| [2] |
|
|
程世平, 姚鹏强, 耿喜宁, 刘春洋, 谢丽华. 2022. 高温诱导牡丹产生未减数花粉. 园艺学报, 49(3):581-589.
|
|
| [3] |
|
| [4] |
|
| [5] |
|
|
康向阳. 2020. 林木三倍体育种研究进展及展望. 中国科学:生命科学, 50(2):136-143.
|
|
| [6] |
|
|
康云艳, 宋爱婷, 柴喜荣, 杨暹. 2022. 黄瓜幼苗茎段石蜡切片制作和番红染色实验方法改进. 实验技术与管理, 39(1):182-184,190.
|
|
| [7] |
|
| [8] |
|
|
李鑫. 2023. 牡丹种子发育及油脂合成相关转录因子分析及鉴定[硕士论文]. 重庆: 重庆师范大学.
|
|
| [9] |
|
| [10] |
|
| [11] |
|
|
刘春洋, 彭朝凤, 程世平, 姚鹏强, 耿喜宁, 谢丽华. 2023. 高温诱导‘凤丹’牡丹2n雌配子创制三倍体. 园艺学报, 50(7):1455-1466.
|
|
| [12] |
|
|
刘慧春, 许雯婷, 周江华, 张加强, 史小华, 朱开元. 2024. 基于牡丹涝害胁迫的转录组分析及SSR引物开发. 浙江农业学报, 36(3):544-558.
|
|
| [13] |
|
| [14] |
|
|
马鸿文, 任宇昕, 龙羿辛, 王楠, 冯祥元, 俞天泉, 华晓琴, 王君. 2024. 青黑杨杂种全同胞二倍体与三倍体长枝叶性状变异研究. 北京林业大学学报, 46(1):27-34.
|
|
| [15] |
|
|
毛常丽, 李玲, 张凤良, 李小琴, 杨湉, 吴裕. 2022. 低温胁迫下巴西橡胶树三倍体全长转录组测序分析. 热带农业科技, 45(1):1-6.
|
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
|
裴芸, 虞夏清, 赵晓坤, 张万萍, 陈劲枫. 2023. 多倍化与植物新表型关联性的研究进展. 园艺学报, 50(9):1854-1866.
|
|
| [21] |
|
| [22] |
|
| [23] |
|
|
王春夏, 尹玥, 王志平, 严瑞, 付麟岚, 孙红梅. 2019. 多倍化兰州百合和细叶百合组培苗再生和耐非生物胁迫能力. 园艺学报, 46(12):2359-2368.
|
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
|
谢洋, 邢雨蒙, 周国彦, 刘美彦, 银珊珊, 闫立英. 2023. 黄瓜二倍体及其同源四倍体果实转录组分析. 生物技术通报, 39(3):152-162.
|
|
| [28] |
|
| [29] |
|
|
杨敏, 龙宇, 王良新, 刘晓阳, 侯国艳, 蒋语嫣, 罗娅. 2022. 胞质甘油醛-3-磷酸脱氢酶GAPC2参与番茄果实成熟的调控. 四川农业大学学报, 40(2):156-162
|
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
|
赵晋锋, 杜艳伟, 余爱丽. 2020. 谷子MDH基因非生物逆境响应特性研究. 核农学报, 34(10):2152-2160.
|
|
| [35] |
|
| [36] |
|
| [1] | LI Ziyan, CHEN Weixi, LI Zihan, LI Yin, LIANG Fengming, ZENG Xiangli, JIAN Hongju, and LÜ Dianqiu. Screening Candidate Genes Controlling Potato Maturation Time Based on RNA-Seq [J]. Acta Horticulturae Sinica, 2025, 52(6): 1505-1518. |
| [2] | FAN Amei, JIN Yaxin, HE Longjiao, LI Qi, ZHANG Yanqing, SHAO Qi, SONG Yun, NIU Yanbing, and QIAO Yonggang. Effects of Gibberellin on the Growth and Development of Forsythia suspensa Heterostyly [J]. Acta Horticulturae Sinica, 2025, 52(11): 3016-3030. |
| [3] | PEI Yun, YU Xiaqing, ZHAO Xiaokun, ZHANG Wanping, CHEN Jinfeng. Progress in the Study of the Association of Polyploidy with New Plant Phenotypes [J]. Acta Horticulturae Sinica, 2023, 50(9): 1854-1866. |
| [4] | LIU Chunyang, PENG Chaofeng, CHENG Shiping, YAO Pengqiang, GENG Xining, XIE Lihua. Creation of Triploid Germplasm in Paeonia ostii‘Fengdan’Through 2n Female Gametes Inducing with High Temperature Treatment [J]. Acta Horticulturae Sinica, 2023, 50(7): 1455-1466. |
| [5] | YUAN Huazhao, PANG Fuhua, WANG Jing, CAI Weijian, XIA Jin, ZHAO Mizhen. Identification of Anthocyanin Compositions and Expression Analysis of Key Related Genes in Fragaria × ananassa [J]. Acta Horticulturae Sinica, 2023, 50(4): 791-801. |
| [6] | ZHU Wei, CAO Jinjin, CHEN Xi, ZHANG Wei, SUN Rongze, ZHU Shaocai, ZHAO Jiageng, CUI Yaqi, WANG Yuxuan, YU Xiaonan. New Herbaceous Peony Cultivars‘Fluttering Pink’‘Fairy’s Cheek’ ‘Blushing Smile’and‘Tiny Lotus’ [J]. Acta Horticulturae Sinica, 2023, 50(2): 457-458. |
| [7] | JIANG Jingdong, WEI Zhuangmin, WANG Nan, ZHU Chenqiao, YE Junli, XIE Zongzhou, DENG Xiuxin, CHAI Lijun. Exploitation and Identification of Tetraploid Resources of Hongkong Kumquat(Fortunella hindsii) [J]. Acta Horticulturae Sinica, 2023, 50(1): 27-35. |
| [8] | ZHANG Wanqing, ZHANG Hongxiao, LIAN Xiaofang, LI Yuying, GUO Lili, HOU Xiaogai. Analysis of DNA Methylation Related to Callus Differentiation and Rooting Induction of Paeonia ostii‘Fengdan’ [J]. Acta Horticulturae Sinica, 2022, 49(8): 1735-1746. |
| [9] | WU Ting, JIA Ruidong, YANG Shuhua, ZHAO Xin, YU Xiaonan, GUO Yuan, GE Hong. Research Advances and Prospects on Phalaenopsis Polyploid Breeding [J]. Acta Horticulturae Sinica, 2022, 49(2): 448-462. |
| [10] | QIAO Jun, WANG Liying, LIU Jing, LI Suweng. Expression Analysis of Genes Related to Photosensitive Color Under the Caylx in Eggplant Based on Transcriptome Sequencing [J]. Acta Horticulturae Sinica, 2022, 49(11): 2347-2356. |
| [11] | JU Lixiang,LEI Xin,ZHAO Chengzhi,SHU Huangying,WANG Zhiwei,and CHENG Shanhan*. Identification of MYB Family Genes and Its Relationship with Pungency of Pepper [J]. ACTA HORTICULTURAE SINICA, 2020, 47(5): 875-892. |
| [12] | CHEN Yao1,2,ZHOU Hanmei1,HE Bing1,*,and LI Wei1. Preliminary Study on the Regulation Mechanism of GA3 and IAA Combination in Paris polyphylla var. chinensis Seed Germination [J]. ACTA HORTICULTURAE SINICA, 2020, 47(2): 321-333. |
| [13] | ZHOU Rui, XIE Kaidong, WANG Wei, PENG Jun, XIE Shanpeng, HU Yibo, WU Xiaomeng, and GUO Wenwu, . Efficient Identification of Tetraploid Plants from Seedling Populations of Apomictic Citrus Genotypes Based on Morphological Characteristics [J]. Acta Horticulturae Sinica, 2020, 47(12): 2451-2458. |
| [14] | DANG Jiangbo1,LIANG Guolu1,LI Cai2,WU Di1,GUO Qigao1,*,LIANG Senlin1,and WANG Peng1. Polyploid Rootstock of Fruit Tree:Research Status and Prospects [J]. ACTA HORTICULTURAE SINICA, 2019, 46(9): 1701-1710. |
| [15] | DONG Yuhui1,WANG Lixia1,GU Qiyu1,LIU Zhongliang2,SUN Xiudong1,and LIU Shiqi1,*. Analysis of Differentially Expressed Genes of Exogenous GA3 Releasing Aerial Bulbs Dormancy in Garlic Based on Transcriptome Sequencing [J]. ACTA HORTICULTURAE SINICA, 2019, 46(12): 2335-2346. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd