
园艺学报 ›› 2024, Vol. 51 ›› Issue (5): 1017-1032.doi: 10.16420/j.issn.0513-353x.2023-0301
袁娜1, 徐勤圆1, 徐照龙1, 周玲1, 刘晓庆2, 陈新2,*( ), 杜建厂1,*(
), 杜建厂1,*( )
)
收稿日期:2023-06-30
修回日期:2023-11-30
出版日期:2024-05-25
发布日期:2024-05-29
通讯作者:
* E-mail:cx@jaas.ac.cn,dujianchang@hotmail.com
基金资助:
YUAN Na1, XU Qinyuan1, XU Zhaolong1, ZHOU Ling1, LIU Xiaoqing2, CHEN Xin2,*( ), DU Jianchang1,*(
), DU Jianchang1,*( )
)
Received:2023-06-30
Revised:2023-11-30
Published:2024-05-25
Online:2024-05-29
Contact:
* E-mail:cx@jaas.ac.cn,dujianchang@hotmail.com
摘要:
根据公开发表的菜豆基因组和重测序数据,利用生物信息学的方法筛选出61 950个高质量SNP位点,通过对目标位点进行探针设计评估,最终设计开发出分别包含40 352、20 855和10 486个SNP位点的40K、20K和10K液相芯片。利用10K液相芯片,对89份菜豆品种进行靶向测序基因分型检测,结果显示菜豆的平均多态性信息含量值和基因多样性值分别为0.35和0.29。STRUCTURE和聚类分析将89份菜豆划分为2个品种群,该分类结果显示同一地域的菜豆品种遗传背景和果实性状大都较为相似。基于选择信号分析,共鉴定到103个处于强烈选择状态的区域,这些区域中的位点可为后续功能基因研究提供候选。
袁娜, 徐勤圆, 徐照龙, 周玲, 刘晓庆, 陈新, 杜建厂. 基于靶向测序技术的菜豆SNP液相芯片开发及验证[J]. 园艺学报, 2024, 51(5): 1017-1032.
YUAN Na, XU Qinyuan, XU Zhaolong, ZHOU Ling, LIU Xiaoqing, CHEN Xin, DU Jianchang. The Development and Verification of SNP Lquid Chips for Common Bean Based on Targeted Sequencing Technology[J]. Acta Horticulturae Sinica, 2024, 51(5): 1017-1032.
| 来源 | 编号 | 品种 | 来源 | 编号 | 品种 |
|---|---|---|---|---|---|
| Origin | Code | Name | Origin | Code | Name |
| 黑龙江省尚志市亚布力致富源良种繁育场Heilongjiang Shangzhi Yabuli Zhifuyuan Stock Breeding Center | Pv01 | 香蕉豆 Xiangjiaodou | 南京绿领种业有限公司 Nanjing Lvling Seed Co.,Ltd | Pv46 | 中华一号 Zhonghua 1 |
| Pv02 | 压趴架大油豆 Yapajia Dayoudou | 南京理想农业科技有限公司 Nanjing Ideal Seed Co.,Ltd | Pv47 | 荚满地 Jiamandi | |
| Pv03 | 一棵松油豆 Yikesong Youdou | 江苏地方品种 Jiangsu Local Cultivar | Pv48 | 19-05叶海黑粒四季豆 19-05 Yehai Heili Sijidou | |
| 黑龙江省尚志市亚布力种子繁育基地 Heilongjiang Shangzhi Yabuli Seed Breeding Base | Pv04 | 兔子翻白眼 Tuzi Fanbaiyan | Pv49 | 1906仪征四季豆 1906 Yizheng Sijidou | |
| 黑龙江亚布力庆丰种业繁育中心 Heilongjiang Yabuli Qingfeng Seed Breeding Center | Pv05 | 开锅烂油豆 Kaiguolan Youdou | 浙江勿忘农种业股份有限公司 Zhejiang Wuwangnong Seeds Shareholding Co.,Ltd | Pv50 | 丽芸3号 Liyun 3 |
| 黑龙江省尚志市亚布力镇蔬菜繁育基地Heilongjiang Shangzhi Yabuli Vegetable Breeding Center | Pv06 | 九月青油豆 Jiuyueqing Youdou | Pv51 | 丽芸2号 Liyun 2 | |
| 黑龙江亚布力蔬菜种子繁育基地 Heilongjiang Yabuli Vegetable Seed Breeding Center | Pv07 | 大马掌油豆 Damazhang Youdou | 杭州科丰种子有限公司 Hangzhou Kefeng Seed Co.,Ltd | Pv52 | 红花青荚四季豆 Honghua Qingjia Sijidou |
| 哈尔滨市金龙农业有限公司 Harbin Jinling Agriculture Co.,Ltd | Pv08 | 日本青刀豆 Ribenqing Daodou | Pv53 | 龙丰一号四季豆 Longfeng 1 Sijidou | |
| Pv09 | 黄金钩地豆 Huangjingou Didou | 浙江之豇种业有限公司 Zhejiang Zhijiang Seed Co.,Ltd | Pv54 | 浙青2号 Zheqing 2 | |
| Pv10 | 勾勾黄架豆 Gougouhuang Jiadou | Pv55 | 浙芸3号 Zheyun 3 | ||
| 哈尔滨市农业科学院 Harbin Academy of Agricultural and Sciences | Pv11 | 绿优早地油豆 Lüyouzao Diyoudou | 上海农乐种植有限公司 Shanghai Nongle Planting Co.,Ltd | Pv56 | 红筋架豆王 Hongjin Jiadouwang |
| 哈尔滨阿城市利农良种研究所 Harbin Acheng Lilongliangzhong Insitute | Pv12 | 三叶紫花架油豆 Sanye Zihua Jiayoudou | Pv57 | 无筋地刀豆 Wujin Didaodou | |
| 吉林省扶余市益农种业有限公司 Jilin Fuyu Yinong Seed Co.,Ltd | Pv13 | 龙泉九粒白 Longquan Jiulibai | 江西宜春市信用种苗中心 Jiangxi Yichun Credit Seedling Centre | Pv58 | 红花四季豆 Honghua Sijidou |
| Pv14 | 八月忙冻死鬼 Bayuemang Dongsi gui | 长沙新万农种业有限公司 Changsha Xinwannong Seed Co.,Ltd | Pv59 | 赛嫩玉豆 Sainen Yudou | |
| 吉林鑫鑫菜籽商店 Jilin Xinxin Canola Store | Pv15 | 将军一点红 Jiangjun Yidianhong | 四川省绵阳市华灵高科良种繁育研究中心Sichun Mianyang Hualing Gaoke Breeding Research Center | Pv60 | 红花白荚 Honghua Baijia |
| Pv16 | 无筋泰国架豆王 Wujin Taiguo Jiadouwang | 四川省绵阳科兴种业有限公司 Sichuan Mianyang Kexing Seed Co.,Ltd | Pv61 | 科兴无筋架豆 Kexing Wujin Jiadou | |
| 吉林盛宇种业有限公司 Jilin Shengyu Seed Co.,Ltd | Pv17 | 抗病大马掌 Kangbing Damazhang | 南宁桂研种业有限公司 Nanning Guiyan Seed Co.,Ltd | Pv62 | 泰优玉美人8号 Taiyou Yumeiren 8 |
| 河北金盛优品农业科技有限公司 Hebei Jinsheng Youpin Agricultural Technology Co.,Ltd | Pv18 | 摘不败八月黄 Zhaibubai Bayuehuang | Pv63 | 绿宝高产36号玉豆 Lübao Gaochan 36 Yudou | |
| 吉林农家品种 Jilin Local Cultivar | Pv19 | 家雀蛋 Jiaquedan | Pv64 | 精选双青十二号玉豆Jingxuan Shuangqing 12 Yudou | |
| 河北青县纯丰蔬菜良种繁育场 Hebei Qing County Chunfeng Vegetable Breeding Center | Pv20 | 雪莲架豆 Xuelian Jiadou | Pv65 | 泰优18号玉豆 Taiyou 18 Yudou | |
| Pv21 | 华煜快豆 Huayukuaidou | Pv66 | 泰丰19号玉豆 Taifeng 19 Yudou | ||
| 沧州津科力丰种苗有限责任公司 Cangzhou Jinke Lifeng Seedling Co.,Ltd | Pv22 | 秋紫豆 Qiuzidou | Pv67 | 泰美春宝32号 Taimei Chunbao 32 | |
| Pv23 | 白不老架豆王 Baibulao Jiadouwang | Pv68 | 泰秀30号玉豆 Taixiu 30 Yudou | ||
| 河北青县纯丰蔬菜良种繁育场 Hebei Qing County Chunfeng Vegetable Breeding Center | Pv24 | 中华红架豆 Zhonghuahong Jiadou | Pv69 | 绿优28号玉豆 Lüyou 28 Yudou | |
| Pv25 | 天马地豆 Tianma Didou | Pv70 | 广泰秀美28号 Guangtai Xiumei 28 | ||
| 河北拖车头农业科技有限公司 Hebei Tuochetou Agricultural Technology Co.,Ltd | Pv26 | 红灯笼 Hongdenglong | Pv71 | 泰玉20号玉豆 Taiyu 20 Yudou | |
| Pv27 | 一代双骄 Yidai Shuangjiao | 广东省江门市新兴种业有限公司Guangdong Jiangmen Xinxing Seed Co.,Ltd | Pv72 | 地豆王二号 Didouwang 2 | |
| Pv28 | 黄金豆 Huangjindou | 海口永丰华生种子有限公司 Haihou Yongfeng Huasheng Seed Co.,Ltd | Pv73 | 广州双青一号玉豆 Guangzhou Shuangqing 1 Yudou | |
| 定州市富胜种子蔬菜销售有限公司 Dingzhou Fusheng Vegetable Seed Co.,Ltd | Pv29 | 北京宽荚白莲 Beijing Kuanjia Bailian | 厦农(厦门)农业科技有限公司Xianong(Xiamen) Agricultural Technology Co.,Ltd | Pv74 | 青芸直尚 Qingyun Zhishang |
| 河北地方品种 Hebei Local Cultivar | Pv30 | 鞍纹大芸豆1 Anwen Dayundou 1 | 未知 Unknown | Pv75 | 大白芸豆 Dabai Yundou |
| Pv31 | 鞍纹大芸豆2 Anwen Dayundou 2 | 青岛农业科学院 Qingdao Academy of Agricultural and Sciences | Pv76 | 2018YD45 | |
| 北京金盛优品农业科技有限公司 Beijing Jinsheng Youpin Agricultural Technology Co.,Ltd | Pv32 | 新无筋早地豆 Xinwujin Zaodidou | Pv77 | 2018YD04 | |
| 天津市蓟宏农业科技有限公司 Tianjing Jihong Agricultural Technology Co.,Ltd | Pv33 | 一挂鞭架豆 Yiguabian Jiadou | 青岛农业科学院 Qingdao Academy of Agricultural and Sciences | Pv78 | 2018YD46 |
| 天津市蓟农种子有限公司 Tianjing Jinong Seed Co.,Ltd | Pv34 | 蓟农双丰二号 Jinong Shuangfeng 2 | Pv79 | 2018YD29 | |
| Pv35 | 阿维斯双丰三号 Aweisi Shuangfeng 3 | Pv80 | 2018YD13 | ||
| 内蒙古农家品种 Inner Mongolia Local Cultivar | Pv36 | 农家红芸豆 Nongjia Hongyundou | Pv81 | 2018YD03 | |
| 新乡市神牛种子有限公司 Xinxiang City Shenniu Seed Co.,Ltd | Pv37 | 摘不败秋紫豆 Zhaibubai Qiuzidou | Pv82 | 2018YD10 | |
| 宁夏平罗兴农蔬菜种苗有限公司 Ningxia Luoping Xingnong Vegetable Seedling Co.,Ltd | Pv38 | 宁兴高产地豆王 Ningxing Gaochan Didouwang | Pv83 | 2018YD26 | |
| 山西地方品种 Shanxi Local Cultivar | Pv39 | 红宝石芸豆 Hongbaoshi Yundou | Pv84 | 2018YD16 | |
| 山东德州市德城区德超种业有限公司 Shandong Dezhou Decheng Dechao Seed Co.,Ltd | Pv40 | 特嫩压塌架 Tenen Yatajia | Pv85 | 2018YD06 | |
| 山东德州市德蔬种业有限公司 Shandong Dezhou Deshu Seed Co.,Ltd | Pv41 | 德蔬地豆王 Deshu Didouwang | Pv86 | 2018YD48 | |
| Pv42 | 红玉天使架豆 Hongyu Tianshi Jiadou | Pv87 | 2018YD01 | ||
| 山东临朐县天和种业有限公司 Shandong Linqu County Tianhe Seed Co.,Ltd | Pv43 | 老来少架豆王 Laolaishao Jiadouwang | Pv88 | 2018YD49 | |
| 莱阳华绿种苗有限公司 Laiyang Hualü Seedling Co.,Ltd | Pv44 | 浓绿江户川青刀豆 Nonglü Jianghuchuan Qingdaodou | Pv89 | 2018YD21 | |
| 济南龙泉种子有限公司 Jinan Longquan Seed Co.,Ltd | Pv45 | 超级九粒白 Chaoji Jiulibai |
表1 菜豆89份品种名称及来源
Table 1 Names and origins of 89 common bean cultivars
| 来源 | 编号 | 品种 | 来源 | 编号 | 品种 |
|---|---|---|---|---|---|
| Origin | Code | Name | Origin | Code | Name |
| 黑龙江省尚志市亚布力致富源良种繁育场Heilongjiang Shangzhi Yabuli Zhifuyuan Stock Breeding Center | Pv01 | 香蕉豆 Xiangjiaodou | 南京绿领种业有限公司 Nanjing Lvling Seed Co.,Ltd | Pv46 | 中华一号 Zhonghua 1 |
| Pv02 | 压趴架大油豆 Yapajia Dayoudou | 南京理想农业科技有限公司 Nanjing Ideal Seed Co.,Ltd | Pv47 | 荚满地 Jiamandi | |
| Pv03 | 一棵松油豆 Yikesong Youdou | 江苏地方品种 Jiangsu Local Cultivar | Pv48 | 19-05叶海黑粒四季豆 19-05 Yehai Heili Sijidou | |
| 黑龙江省尚志市亚布力种子繁育基地 Heilongjiang Shangzhi Yabuli Seed Breeding Base | Pv04 | 兔子翻白眼 Tuzi Fanbaiyan | Pv49 | 1906仪征四季豆 1906 Yizheng Sijidou | |
| 黑龙江亚布力庆丰种业繁育中心 Heilongjiang Yabuli Qingfeng Seed Breeding Center | Pv05 | 开锅烂油豆 Kaiguolan Youdou | 浙江勿忘农种业股份有限公司 Zhejiang Wuwangnong Seeds Shareholding Co.,Ltd | Pv50 | 丽芸3号 Liyun 3 |
| 黑龙江省尚志市亚布力镇蔬菜繁育基地Heilongjiang Shangzhi Yabuli Vegetable Breeding Center | Pv06 | 九月青油豆 Jiuyueqing Youdou | Pv51 | 丽芸2号 Liyun 2 | |
| 黑龙江亚布力蔬菜种子繁育基地 Heilongjiang Yabuli Vegetable Seed Breeding Center | Pv07 | 大马掌油豆 Damazhang Youdou | 杭州科丰种子有限公司 Hangzhou Kefeng Seed Co.,Ltd | Pv52 | 红花青荚四季豆 Honghua Qingjia Sijidou |
| 哈尔滨市金龙农业有限公司 Harbin Jinling Agriculture Co.,Ltd | Pv08 | 日本青刀豆 Ribenqing Daodou | Pv53 | 龙丰一号四季豆 Longfeng 1 Sijidou | |
| Pv09 | 黄金钩地豆 Huangjingou Didou | 浙江之豇种业有限公司 Zhejiang Zhijiang Seed Co.,Ltd | Pv54 | 浙青2号 Zheqing 2 | |
| Pv10 | 勾勾黄架豆 Gougouhuang Jiadou | Pv55 | 浙芸3号 Zheyun 3 | ||
| 哈尔滨市农业科学院 Harbin Academy of Agricultural and Sciences | Pv11 | 绿优早地油豆 Lüyouzao Diyoudou | 上海农乐种植有限公司 Shanghai Nongle Planting Co.,Ltd | Pv56 | 红筋架豆王 Hongjin Jiadouwang |
| 哈尔滨阿城市利农良种研究所 Harbin Acheng Lilongliangzhong Insitute | Pv12 | 三叶紫花架油豆 Sanye Zihua Jiayoudou | Pv57 | 无筋地刀豆 Wujin Didaodou | |
| 吉林省扶余市益农种业有限公司 Jilin Fuyu Yinong Seed Co.,Ltd | Pv13 | 龙泉九粒白 Longquan Jiulibai | 江西宜春市信用种苗中心 Jiangxi Yichun Credit Seedling Centre | Pv58 | 红花四季豆 Honghua Sijidou |
| Pv14 | 八月忙冻死鬼 Bayuemang Dongsi gui | 长沙新万农种业有限公司 Changsha Xinwannong Seed Co.,Ltd | Pv59 | 赛嫩玉豆 Sainen Yudou | |
| 吉林鑫鑫菜籽商店 Jilin Xinxin Canola Store | Pv15 | 将军一点红 Jiangjun Yidianhong | 四川省绵阳市华灵高科良种繁育研究中心Sichun Mianyang Hualing Gaoke Breeding Research Center | Pv60 | 红花白荚 Honghua Baijia |
| Pv16 | 无筋泰国架豆王 Wujin Taiguo Jiadouwang | 四川省绵阳科兴种业有限公司 Sichuan Mianyang Kexing Seed Co.,Ltd | Pv61 | 科兴无筋架豆 Kexing Wujin Jiadou | |
| 吉林盛宇种业有限公司 Jilin Shengyu Seed Co.,Ltd | Pv17 | 抗病大马掌 Kangbing Damazhang | 南宁桂研种业有限公司 Nanning Guiyan Seed Co.,Ltd | Pv62 | 泰优玉美人8号 Taiyou Yumeiren 8 |
| 河北金盛优品农业科技有限公司 Hebei Jinsheng Youpin Agricultural Technology Co.,Ltd | Pv18 | 摘不败八月黄 Zhaibubai Bayuehuang | Pv63 | 绿宝高产36号玉豆 Lübao Gaochan 36 Yudou | |
| 吉林农家品种 Jilin Local Cultivar | Pv19 | 家雀蛋 Jiaquedan | Pv64 | 精选双青十二号玉豆Jingxuan Shuangqing 12 Yudou | |
| 河北青县纯丰蔬菜良种繁育场 Hebei Qing County Chunfeng Vegetable Breeding Center | Pv20 | 雪莲架豆 Xuelian Jiadou | Pv65 | 泰优18号玉豆 Taiyou 18 Yudou | |
| Pv21 | 华煜快豆 Huayukuaidou | Pv66 | 泰丰19号玉豆 Taifeng 19 Yudou | ||
| 沧州津科力丰种苗有限责任公司 Cangzhou Jinke Lifeng Seedling Co.,Ltd | Pv22 | 秋紫豆 Qiuzidou | Pv67 | 泰美春宝32号 Taimei Chunbao 32 | |
| Pv23 | 白不老架豆王 Baibulao Jiadouwang | Pv68 | 泰秀30号玉豆 Taixiu 30 Yudou | ||
| 河北青县纯丰蔬菜良种繁育场 Hebei Qing County Chunfeng Vegetable Breeding Center | Pv24 | 中华红架豆 Zhonghuahong Jiadou | Pv69 | 绿优28号玉豆 Lüyou 28 Yudou | |
| Pv25 | 天马地豆 Tianma Didou | Pv70 | 广泰秀美28号 Guangtai Xiumei 28 | ||
| 河北拖车头农业科技有限公司 Hebei Tuochetou Agricultural Technology Co.,Ltd | Pv26 | 红灯笼 Hongdenglong | Pv71 | 泰玉20号玉豆 Taiyu 20 Yudou | |
| Pv27 | 一代双骄 Yidai Shuangjiao | 广东省江门市新兴种业有限公司Guangdong Jiangmen Xinxing Seed Co.,Ltd | Pv72 | 地豆王二号 Didouwang 2 | |
| Pv28 | 黄金豆 Huangjindou | 海口永丰华生种子有限公司 Haihou Yongfeng Huasheng Seed Co.,Ltd | Pv73 | 广州双青一号玉豆 Guangzhou Shuangqing 1 Yudou | |
| 定州市富胜种子蔬菜销售有限公司 Dingzhou Fusheng Vegetable Seed Co.,Ltd | Pv29 | 北京宽荚白莲 Beijing Kuanjia Bailian | 厦农(厦门)农业科技有限公司Xianong(Xiamen) Agricultural Technology Co.,Ltd | Pv74 | 青芸直尚 Qingyun Zhishang |
| 河北地方品种 Hebei Local Cultivar | Pv30 | 鞍纹大芸豆1 Anwen Dayundou 1 | 未知 Unknown | Pv75 | 大白芸豆 Dabai Yundou |
| Pv31 | 鞍纹大芸豆2 Anwen Dayundou 2 | 青岛农业科学院 Qingdao Academy of Agricultural and Sciences | Pv76 | 2018YD45 | |
| 北京金盛优品农业科技有限公司 Beijing Jinsheng Youpin Agricultural Technology Co.,Ltd | Pv32 | 新无筋早地豆 Xinwujin Zaodidou | Pv77 | 2018YD04 | |
| 天津市蓟宏农业科技有限公司 Tianjing Jihong Agricultural Technology Co.,Ltd | Pv33 | 一挂鞭架豆 Yiguabian Jiadou | 青岛农业科学院 Qingdao Academy of Agricultural and Sciences | Pv78 | 2018YD46 |
| 天津市蓟农种子有限公司 Tianjing Jinong Seed Co.,Ltd | Pv34 | 蓟农双丰二号 Jinong Shuangfeng 2 | Pv79 | 2018YD29 | |
| Pv35 | 阿维斯双丰三号 Aweisi Shuangfeng 3 | Pv80 | 2018YD13 | ||
| 内蒙古农家品种 Inner Mongolia Local Cultivar | Pv36 | 农家红芸豆 Nongjia Hongyundou | Pv81 | 2018YD03 | |
| 新乡市神牛种子有限公司 Xinxiang City Shenniu Seed Co.,Ltd | Pv37 | 摘不败秋紫豆 Zhaibubai Qiuzidou | Pv82 | 2018YD10 | |
| 宁夏平罗兴农蔬菜种苗有限公司 Ningxia Luoping Xingnong Vegetable Seedling Co.,Ltd | Pv38 | 宁兴高产地豆王 Ningxing Gaochan Didouwang | Pv83 | 2018YD26 | |
| 山西地方品种 Shanxi Local Cultivar | Pv39 | 红宝石芸豆 Hongbaoshi Yundou | Pv84 | 2018YD16 | |
| 山东德州市德城区德超种业有限公司 Shandong Dezhou Decheng Dechao Seed Co.,Ltd | Pv40 | 特嫩压塌架 Tenen Yatajia | Pv85 | 2018YD06 | |
| 山东德州市德蔬种业有限公司 Shandong Dezhou Deshu Seed Co.,Ltd | Pv41 | 德蔬地豆王 Deshu Didouwang | Pv86 | 2018YD48 | |
| Pv42 | 红玉天使架豆 Hongyu Tianshi Jiadou | Pv87 | 2018YD01 | ||
| 山东临朐县天和种业有限公司 Shandong Linqu County Tianhe Seed Co.,Ltd | Pv43 | 老来少架豆王 Laolaishao Jiadouwang | Pv88 | 2018YD49 | |
| 莱阳华绿种苗有限公司 Laiyang Hualü Seedling Co.,Ltd | Pv44 | 浓绿江户川青刀豆 Nonglü Jianghuchuan Qingdaodou | Pv89 | 2018YD21 | |
| 济南龙泉种子有限公司 Jinan Longquan Seed Co.,Ltd | Pv45 | 超级九粒白 Chaoji Jiulibai |
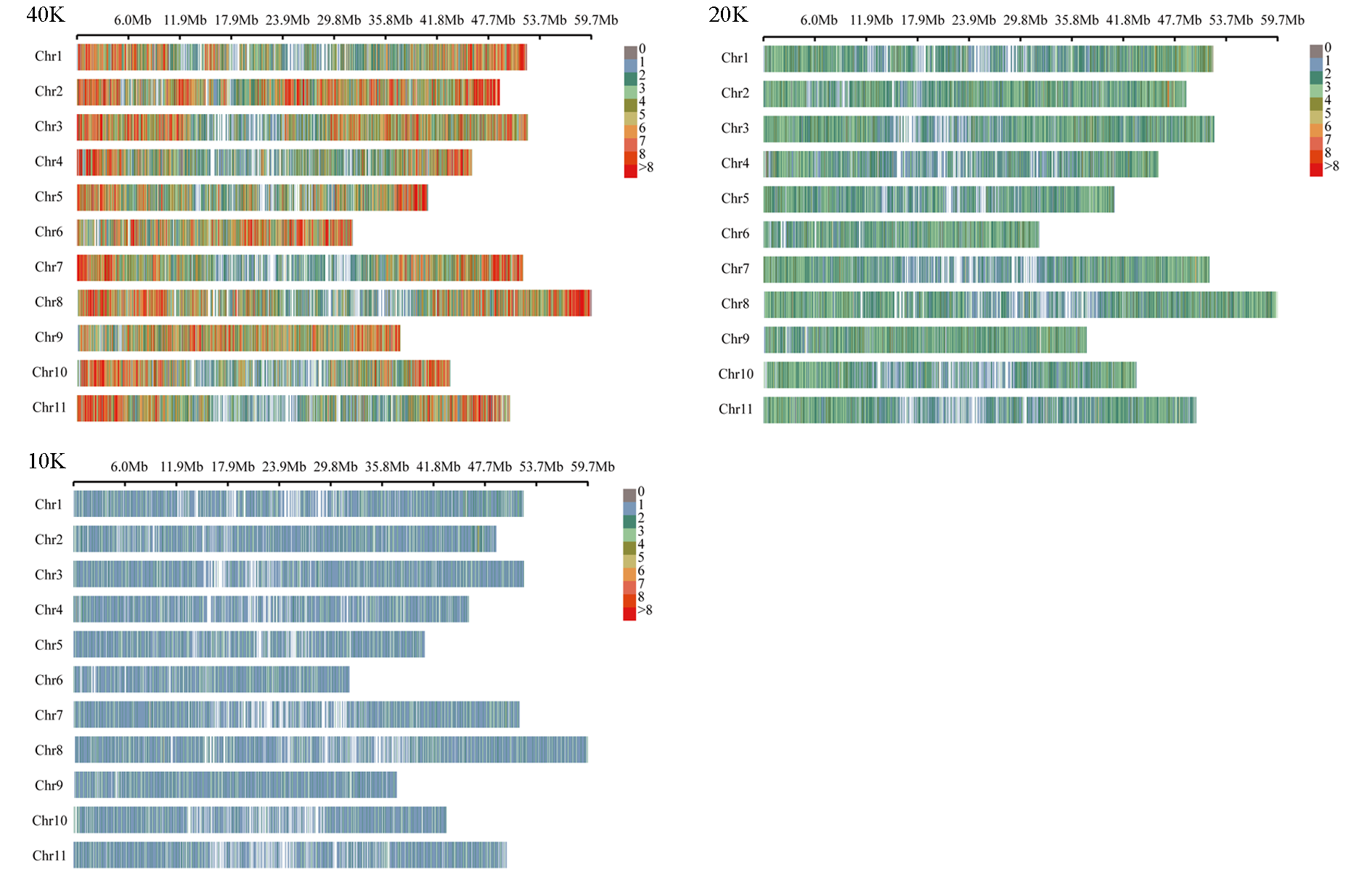
图1 40K、20K和10K芯片面板中SNP标记在菜豆11条染色体上的分布模式 标记密度由不同柱形颜色表示,每个柱形代表1 Mb窗口大小。
Fig. 1 Distribution of SNP markers on 11 common bean chromosomes for three marker panels The marker density is indicated by different barcolors,and each bar represents 1 Mb window size.

图3 40K、20K和10K芯片面板中的SNP类型(A)、最小等位基因频率(B)、各染色体上SNP数量(C)及基因多样性和多态性信息含量(D)
Fig. 3 SNP type(A),minor allele frequency(B),number of SNPs on each chromosome(C),gene diversity and polymorphism information content(D)in the three marker panels
| 编号 Code | 原始读长数 Raw reads | 干净读长数 Clean reads | 原始碱基数/bp Raw bases | 干净碱基数/bp Clean bases | 比对读长数 Align reads | 比对率/% Align rate | 有效率/% Effective rate | Q20 | Q30 |
|---|---|---|---|---|---|---|---|---|---|
| Pv01 | 8 098 852 | 8 070 766 | 1 214 827 800 | 1 173 728 992 | 7 656 019 | 94.86 | 96.62 | 97.74 | 92.44 |
| Pv02 | 5 789 802 | 5 770 124 | 868 470 300 | 825 811 078 | 5 359 340 | 92.88 | 95.09 | 98.01 | 93.42 |
| Pv03 | 7 816 136 | 7 789 678 | 1 172 420 400 | 1 131 921 434 | 7 323 176 | 94.01 | 96.55 | 97.45 | 91.61 |
| Pv04 | 7 047 902 | 7 024 020 | 1 057 185 300 | 1 011 814 446 | 6 687 162 | 95.20 | 95.71 | 97.72 | 92.37 |
| Pv05 | 7 080 946 | 7 056 892 | 1 062 141 900 | 1 021 913 214 | 6 685 993 | 94.74 | 96.21 | 97.81 | 92.79 |
| Pv06 | 8 536 526 | 8 508 450 | 1 280 478 900 | 1 226 760 962 | 8 018 564 | 94.24 | 95.80 | 97.79 | 92.63 |
| Pv07 | 5 320 770 | 5 302 078 | 798 115 500 | 767 339 022 | 4 984 408 | 94.01 | 96.14 | 97.88 | 93.06 |
| Pv08 | 5 731 130 | 5 711 446 | 859 669 500 | 818 679 944 | 5 321 662 | 93.18 | 95.23 | 97.76 | 92.49 |
| Pv09 | 8 312 168 | 8 284 078 | 1 246 825 200 | 1 192 209 370 | 7 847 519 | 94.73 | 95.62 | 97.72 | 92.43 |
| Pv10 | 5 737 552 | 5 717 664 | 860 632 800 | 835 172 092 | 5 416 408 | 94.73 | 97.04 | 97.72 | 92.43 |
| Pv11 | 7 636 864 | 7 611 082 | 1 145 529 600 | 1 095 790 714 | 7 164 073 | 94.13 | 95.66 | 97.84 | 92.87 |
| Pv12 | 3 098 470 | 3 087 418 | 464 770 500 | 439 198 858 | 2 853 437 | 92.42 | 94.50 | 98.37 | 94.59 |
| Pv13 | 6 089 206 | 6 068 534 | 913 380 900 | 876 023 234 | 5 525 860 | 91.06 | 95.91 | 97.75 | 92.50 |
| Pv14 | 6 742 030 | 6 719 166 | 1 011 304 500 | 972 387 718 | 6 158 895 | 91.66 | 96.15 | 97.80 | 92.66 |
| Pv15 | 7 133 440 | 7 109 086 | 1 070 016 000 | 1 025 095 922 | 6 667 344 | 93.79 | 95.80 | 97.98 | 93.24 |
| Pv16 | 6 983 360 | 6 959 954 | 1 047 504 000 | 1 004 470 008 | 6 414 434 | 92.16 | 95.89 | 97.55 | 91.89 |
| Pv17 | 4 437 716 | 4 422 562 | 665 657 400 | 633 980 678 | 4 136 234 | 93.53 | 95.24 | 98.26 | 94.22 |
| Pv18 | 5 119 318 | 5 101 940 | 767 897 700 | 726 250 480 | 4 813 235 | 94.34 | 94.58 | 98.15 | 93.88 |
| Pv19 | 7 449 680 | 7 424 586 | 1 117 452 000 | 1 067 824 744 | 7 011 514 | 94.44 | 95.56 | 97.97 | 93.38 |
| Pv20 | 7 918 080 | 7 891 088 | 1 187 712 000 | 1 148 448 130 | 7 223 517 | 91.54 | 96.69 | 97.50 | 91.67 |
| Pv21 | 7 008 252 | 6 984 480 | 1 051 237 800 | 1 014 563 204 | 6 406 385 | 91.72 | 96.51 | 97.57 | 91.90 |
| Pv22 | 4 030 630 | 4 017 024 | 604 594 500 | 579 570 558 | 3 679 759 | 91.60 | 95.86 | 97.52 | 91.81 |
| Pv23 | 5 611 150 | 5 592 006 | 841 672 500 | 809 023 072 | 5 125 314 | 91.65 | 96.12 | 97.28 | 91.15 |
| Pv24 | 3 049 720 | 3 041 280 | 457 458 000 | 392 415 760 | 2 738 569 | 90.05 | 85.78 | 98.58 | 95.24 |
| Pv25 | 4 124 312 | 4 110 136 | 618 646 800 | 589 331 084 | 3 834 557 | 93.30 | 95.26 | 97.80 | 92.68 |
| Pv26 | 5 852 764 | 5 832 946 | 877 914 600 | 849 599 058 | 5 332 649 | 91.42 | 96.77 | 97.69 | 92.25 |
| Pv27 | 7 049 974 | 7 025 392 | 1 057 496 100 | 1 009 922 856 | 6 460 748 | 91.96 | 95.50 | 97.86 | 92.79 |
| Pv28 | 7 709 688 | 7 683 630 | 1 156 453 200 | 1 106 745 810 | 7 022 063 | 91.39 | 95.70 | 97.60 | 92.04 |
| Pv29 | 7 568 282 | 7 542 736 | 1 135 242 300 | 1 088 137 392 | 6 988 620 | 92.65 | 95.85 | 97.67 | 92.24 |
| Pv30 | 3 002 474 | 2 991 998 | 450 371 100 | 423 707 654 | 2 804 127 | 93.72 | 94.08 | 98.41 | 94.70 |
| Pv31 | 7 769 006 | 7 743 504 | 1 165 350 900 | 1 112 068 434 | 7 347 421 | 94.88 | 95.43 | 97.82 | 92.78 |
| Pv32 | 6 583 480 | 6 562 116 | 987 522 000 | 946 078 096 | 6 143 616 | 93.62 | 95.80 | 97.60 | 92.07 |
| Pv33 | 6 943 390 | 6 919 310 | 1 041 508 500 | 997 557 318 | 6 325 013 | 91.41 | 95.78 | 97.88 | 92.96 |
| Pv34 | 6 651 502 | 6 628 502 | 997 725 300 | 954 889 330 | 6 037 645 | 91.09 | 95.71 | 97.66 | 92.29 |
| Pv35 | 16 515 772 | 16 486 982 | 2 477 365 800 | 2 314 193 540 | 15 197 899 | 92.18 | 93.41 | 98.34 | 94.34 |
| Pv36 | 8 663 510 | 8 634 920 | 1 299 526 500 | 1 235 250 830 | 8 140 449 | 94.27 | 95.05 | 97.63 | 92.14 |
| Pv37 | 6 477 232 | 6 455 580 | 971 584 800 | 927 022 220 | 5 884 989 | 91.16 | 95.41 | 97.77 | 92.73 |
| Pv38 | 8 044 132 | 8 016 870 | 1 206 619 800 | 1 150 362 636 | 7 474 688 | 93.24 | 95.34 | 97.70 | 92.41 |
| Pv39 | 7 639 474 | 7 613 382 | 1 145 921 100 | 1 090 946 618 | 7 174 002 | 94.23 | 95.20 | 97.83 | 92.78 |
| Pv40 | 7 361 614 | 7 337 070 | 1 104 242 100 | 1 066 060 554 | 6 703 574 | 91.37 | 96.54 | 97.57 | 91.86 |
| Pv41 | 7 505 308 | 7 480 256 | 1 125 796 200 | 1 080 668 216 | 6 893 819 | 92.16 | 95.99 | 97.72 | 92.44 |
| Pv42 | 7 348 356 | 7 324 392 | 1 102 253 400 | 1 063 679 164 | 6 768 694 | 92.41 | 96.50 | 97.30 | 91.03 |
| Pv43 | 6 587 980 | 6 565 632 | 988 197 000 | 946 190 986 | 6 004 408 | 91.45 | 95.75 | 97.54 | 91.90 |
| Pv44 | 7 403 192 | 7 379 070 | 1 110 478 800 | 1 058 345 682 | 6 901 485 | 93.53 | 95.31 | 97.57 | 91.97 |
| Pv45 | 7 764 072 | 7 737 444 | 1 164 610 800 | 1 108 132 120 | 7 056 881 | 91.20 | 95.15 | 97.79 | 92.62 |
| Pv46 | 4 830 616 | 4 814 580 | 724 592 400 | 696 981 244 | 4 397 219 | 91.33 | 96.19 | 97.88 | 93.05 |
| Pv47 | 7 547 974 | 7 522 116 | 1 132 196 100 | 1 078 148 092 | 7 025 146 | 93.39 | 95.23 | 97.88 | 92.86 |
| Pv48 | 6 978 052 | 6 954 442 | 1 046 707 800 | 1 001 538 926 | 6 352 621 | 91.35 | 95.68 | 97.79 | 92.65 |
| Pv49 | 6 826 836 | 6 804 008 | 1 024 025 400 | 985 507 780 | 6 214 244 | 91.33 | 96.24 | 97.76 | 92.53 |
| Pv50 | 5 268 676 | 5 251 028 | 790 301 400 | 754 087 146 | 4 785 144 | 91.13 | 95.42 | 97.75 | 92.40 |
| Pv51 | 6 257 576 | 6 236 592 | 938 636 400 | 894 014 262 | 5 688 008 | 91.20 | 95.25 | 97.81 | 92.69 |
| Pv52 | 6 214 442 | 6 193 152 | 932 166 300 | 895 366 264 | 5 662 541 | 91.43 | 96.05 | 97.83 | 92.81 |
| Pv53 | 7 123 026 | 7 099 578 | 1 068 453 900 | 1 025 234 062 | 6 483 231 | 91.32 | 95.95 | 97.67 | 92.23 |
| Pv54 | 2 996 190 | 2 985 946 | 449 428 500 | 430 855 692 | 2 736 528 | 91.65 | 95.87 | 98.09 | 93.66 |
| Pv55 | 3 334 436 | 3 323 366 | 500 165 400 | 482 901 568 | 3 040 272 | 91.48 | 96.55 | 97.29 | 91.07 |
| Pv56 | 5 981 750 | 5 961 578 | 897 262 500 | 864 283 148 | 5 454 974 | 91.50 | 96.32 | 97.51 | 91.69 |
| Pv57 | 6 743 544 | 6 720 248 | 1 011 531 600 | 966 479 926 | 6 292 623 | 93.64 | 95.55 | 97.99 | 93.32 |
| Pv58 | 6 757 314 | 6 734 582 | 1 013 597 100 | 967 411 196 | 6 149 238 | 91.31 | 95.44 | 97.66 | 92.26 |
| Pv59 | 6 644 366 | 6 622 624 | 996 654 900 | 952 675 344 | 6 039 136 | 91.19 | 95.59 | 97.71 | 92.41 |
| Pv60 | 3 308 738 | 3 297 088 | 496 310 700 | 473 938 530 | 3 006 470 | 91.19 | 95.49 | 98.13 | 93.75 |
| Pv61 | 7 164 518 | 7 140 300 | 1 074 677 700 | 1 028 969 692 | 6 606 890 | 92.53 | 95.75 | 97.82 | 92.79 |
| Pv62 | 6 544 594 | 6 522 146 | 981 689 100 | 937 859 972 | 5 945 974 | 91.17 | 95.54 | 97.57 | 91.96 |
| Pv63 | 6 262 790 | 6 242 050 | 939 418 500 | 901 268 214 | 5 721 647 | 91.66 | 95.94 | 97.61 | 92.12 |
| Pv64 | 7 756 110 | 7 729 588 | 1 163 416 500 | 1 110 586 330 | 7 062 279 | 91.37 | 95.46 | 97.69 | 92.31 |
| Pv65 | 7 212 746 | 7 189 088 | 1 081 911 900 | 1 038 137 070 | 6 569 530 | 91.38 | 95.95 | 97.64 | 92.24 |
| Pv66 | 6 842 464 | 6 819 456 | 1 026 369 600 | 980 771 280 | 6 227 660 | 91.32 | 95.56 | 97.80 | 92.66 |
| Pv67 | 5 491 250 | 5 472 356 | 823 687 500 | 786 889 766 | 5 007 569 | 91.51 | 95.53 | 98.22 | 94.03 |
| Pv68 | 8 598 260 | 8 569 182 | 1 289 739 000 | 1 225 043 812 | 7 824 318 | 91.31 | 94.98 | 97.96 | 93.23 |
| Pv69 | 7 098 136 | 7 074 072 | 1 064 720 400 | 1 021 245 826 | 6 476 276 | 91.55 | 95.92 | 97.89 | 92.98 |
| Pv70 | 6 744 486 | 6 721 580 | 1 011 672 900 | 970 160 098 | 6 153 450 | 91.55 | 95.90 | 97.50 | 91.86 |
| Pv71 | 6 954 908 | 6 932 014 | 1 043 236 200 | 997 598 058 | 6 333 065 | 91.36 | 95.63 | 97.56 | 91.86 |
| Pv72 | 6 947 998 | 6 924 592 | 1 042 199 700 | 995 918 234 | 6 390 351 | 92.28 | 95.56 | 97.92 | 93.06 |
| Pv73 | 11 385 340 | 11 365 232 | 1 707 801 000 | 1 521 977 790 | 10 427 776 | 91.75 | 89.12 | 98.37 | 94.62 |
| Pv74 | 6 704 026 | 6 681 088 | 1 005 603 900 | 966 719 208 | 6 098 771 | 91.28 | 96.13 | 97.64 | 92.13 |
| Pv75 | 15 259 508 | 15 229 100 | 2 288 926 200 | 2 115 314 516 | 13 656 417 | 89.67 | 92.42 | 98.24 | 94.14 |
| Pv76 | 6 308 324 | 6 287 108 | 946 248 600 | 904 804 666 | 5 822 194 | 92.61 | 95.62 | 98.04 | 93.56 |
| Pv77 | 5 934 858 | 5 915 132 | 890 228 700 | 860 418 210 | 5 605 331 | 94.76 | 96.65 | 97.57 | 91.79 |
| Pv78 | 7 053 330 | 7 029 220 | 1 057 999 500 | 1 014 776 334 | 6 427 694 | 91.44 | 95.91 | 97.83 | 92.76 |
| Pv79 | 6 626 144 | 6 603 632 | 993 921 600 | 954 116 282 | 6 234 280 | 94.41 | 96.00 | 97.74 | 92.42 |
| Pv80 | 7 477 584 | 7 452 982 | 1 121 637 600 | 1 073 606 700 | 6 968 107 | 93.49 | 95.72 | 97.59 | 92.08 |
| Pv81 | 6 965 426 | 6 941 310 | 1 044 813 900 | 1 002 425 192 | 6 527 168 | 94.03 | 95.94 | 98.23 | 94.06 |
| Pv82 | 7 585 280 | 7 559 696 | 1 137 792 000 | 1 094 479 824 | 6 903 582 | 91.32 | 96.19 | 97.73 | 92.43 |
| Pv83 | 8 660 626 | 8 630 762 | 1 299 093 900 | 1 241 602 578 | 7 908 789 | 91.63 | 95.57 | 97.98 | 93.28 |
| Pv84 | 7 290 040 | 7 264 834 | 1 093 506 000 | 1 048 919 858 | 6 648 774 | 91.52 | 95.92 | 97.60 | 92.08 |
| Pv85 | 7 969 190 | 7 941 526 | 1 195 378 500 | 1 149 028 462 | 7 385 800 | 93.00 | 96.12 | 97.99 | 93.34 |
| Pv86 | 7 584 448 | 7 558 542 | 1 137 667 200 | 1 082 746 918 | 6 911 071 | 91.43 | 95.17 | 98.27 | 94.17 |
| Pv87 | 7 584 774 | 7 558 902 | 1 137 716 100 | 1 083 325 126 | 6 884 039 | 91.07 | 95.22 | 97.91 | 93.08 |
| Pv88 | 7 352 392 | 7 327 872 | 1 102 858 800 | 1 066 357 808 | 6 738 930 | 91.96 | 96.69 | 97.38 | 91.41 |
| Pv89 | 6 766 290 | 6 743 796 | 1 014 943 500 | 967 162 154 | 6 191 315 | 91.81 | 95.29 | 97.74 | 92.52 |
表2 菜豆89个品种的测序数据统计表
Table 2 Statistics on target sequencing of 89 common bean cultivars
| 编号 Code | 原始读长数 Raw reads | 干净读长数 Clean reads | 原始碱基数/bp Raw bases | 干净碱基数/bp Clean bases | 比对读长数 Align reads | 比对率/% Align rate | 有效率/% Effective rate | Q20 | Q30 |
|---|---|---|---|---|---|---|---|---|---|
| Pv01 | 8 098 852 | 8 070 766 | 1 214 827 800 | 1 173 728 992 | 7 656 019 | 94.86 | 96.62 | 97.74 | 92.44 |
| Pv02 | 5 789 802 | 5 770 124 | 868 470 300 | 825 811 078 | 5 359 340 | 92.88 | 95.09 | 98.01 | 93.42 |
| Pv03 | 7 816 136 | 7 789 678 | 1 172 420 400 | 1 131 921 434 | 7 323 176 | 94.01 | 96.55 | 97.45 | 91.61 |
| Pv04 | 7 047 902 | 7 024 020 | 1 057 185 300 | 1 011 814 446 | 6 687 162 | 95.20 | 95.71 | 97.72 | 92.37 |
| Pv05 | 7 080 946 | 7 056 892 | 1 062 141 900 | 1 021 913 214 | 6 685 993 | 94.74 | 96.21 | 97.81 | 92.79 |
| Pv06 | 8 536 526 | 8 508 450 | 1 280 478 900 | 1 226 760 962 | 8 018 564 | 94.24 | 95.80 | 97.79 | 92.63 |
| Pv07 | 5 320 770 | 5 302 078 | 798 115 500 | 767 339 022 | 4 984 408 | 94.01 | 96.14 | 97.88 | 93.06 |
| Pv08 | 5 731 130 | 5 711 446 | 859 669 500 | 818 679 944 | 5 321 662 | 93.18 | 95.23 | 97.76 | 92.49 |
| Pv09 | 8 312 168 | 8 284 078 | 1 246 825 200 | 1 192 209 370 | 7 847 519 | 94.73 | 95.62 | 97.72 | 92.43 |
| Pv10 | 5 737 552 | 5 717 664 | 860 632 800 | 835 172 092 | 5 416 408 | 94.73 | 97.04 | 97.72 | 92.43 |
| Pv11 | 7 636 864 | 7 611 082 | 1 145 529 600 | 1 095 790 714 | 7 164 073 | 94.13 | 95.66 | 97.84 | 92.87 |
| Pv12 | 3 098 470 | 3 087 418 | 464 770 500 | 439 198 858 | 2 853 437 | 92.42 | 94.50 | 98.37 | 94.59 |
| Pv13 | 6 089 206 | 6 068 534 | 913 380 900 | 876 023 234 | 5 525 860 | 91.06 | 95.91 | 97.75 | 92.50 |
| Pv14 | 6 742 030 | 6 719 166 | 1 011 304 500 | 972 387 718 | 6 158 895 | 91.66 | 96.15 | 97.80 | 92.66 |
| Pv15 | 7 133 440 | 7 109 086 | 1 070 016 000 | 1 025 095 922 | 6 667 344 | 93.79 | 95.80 | 97.98 | 93.24 |
| Pv16 | 6 983 360 | 6 959 954 | 1 047 504 000 | 1 004 470 008 | 6 414 434 | 92.16 | 95.89 | 97.55 | 91.89 |
| Pv17 | 4 437 716 | 4 422 562 | 665 657 400 | 633 980 678 | 4 136 234 | 93.53 | 95.24 | 98.26 | 94.22 |
| Pv18 | 5 119 318 | 5 101 940 | 767 897 700 | 726 250 480 | 4 813 235 | 94.34 | 94.58 | 98.15 | 93.88 |
| Pv19 | 7 449 680 | 7 424 586 | 1 117 452 000 | 1 067 824 744 | 7 011 514 | 94.44 | 95.56 | 97.97 | 93.38 |
| Pv20 | 7 918 080 | 7 891 088 | 1 187 712 000 | 1 148 448 130 | 7 223 517 | 91.54 | 96.69 | 97.50 | 91.67 |
| Pv21 | 7 008 252 | 6 984 480 | 1 051 237 800 | 1 014 563 204 | 6 406 385 | 91.72 | 96.51 | 97.57 | 91.90 |
| Pv22 | 4 030 630 | 4 017 024 | 604 594 500 | 579 570 558 | 3 679 759 | 91.60 | 95.86 | 97.52 | 91.81 |
| Pv23 | 5 611 150 | 5 592 006 | 841 672 500 | 809 023 072 | 5 125 314 | 91.65 | 96.12 | 97.28 | 91.15 |
| Pv24 | 3 049 720 | 3 041 280 | 457 458 000 | 392 415 760 | 2 738 569 | 90.05 | 85.78 | 98.58 | 95.24 |
| Pv25 | 4 124 312 | 4 110 136 | 618 646 800 | 589 331 084 | 3 834 557 | 93.30 | 95.26 | 97.80 | 92.68 |
| Pv26 | 5 852 764 | 5 832 946 | 877 914 600 | 849 599 058 | 5 332 649 | 91.42 | 96.77 | 97.69 | 92.25 |
| Pv27 | 7 049 974 | 7 025 392 | 1 057 496 100 | 1 009 922 856 | 6 460 748 | 91.96 | 95.50 | 97.86 | 92.79 |
| Pv28 | 7 709 688 | 7 683 630 | 1 156 453 200 | 1 106 745 810 | 7 022 063 | 91.39 | 95.70 | 97.60 | 92.04 |
| Pv29 | 7 568 282 | 7 542 736 | 1 135 242 300 | 1 088 137 392 | 6 988 620 | 92.65 | 95.85 | 97.67 | 92.24 |
| Pv30 | 3 002 474 | 2 991 998 | 450 371 100 | 423 707 654 | 2 804 127 | 93.72 | 94.08 | 98.41 | 94.70 |
| Pv31 | 7 769 006 | 7 743 504 | 1 165 350 900 | 1 112 068 434 | 7 347 421 | 94.88 | 95.43 | 97.82 | 92.78 |
| Pv32 | 6 583 480 | 6 562 116 | 987 522 000 | 946 078 096 | 6 143 616 | 93.62 | 95.80 | 97.60 | 92.07 |
| Pv33 | 6 943 390 | 6 919 310 | 1 041 508 500 | 997 557 318 | 6 325 013 | 91.41 | 95.78 | 97.88 | 92.96 |
| Pv34 | 6 651 502 | 6 628 502 | 997 725 300 | 954 889 330 | 6 037 645 | 91.09 | 95.71 | 97.66 | 92.29 |
| Pv35 | 16 515 772 | 16 486 982 | 2 477 365 800 | 2 314 193 540 | 15 197 899 | 92.18 | 93.41 | 98.34 | 94.34 |
| Pv36 | 8 663 510 | 8 634 920 | 1 299 526 500 | 1 235 250 830 | 8 140 449 | 94.27 | 95.05 | 97.63 | 92.14 |
| Pv37 | 6 477 232 | 6 455 580 | 971 584 800 | 927 022 220 | 5 884 989 | 91.16 | 95.41 | 97.77 | 92.73 |
| Pv38 | 8 044 132 | 8 016 870 | 1 206 619 800 | 1 150 362 636 | 7 474 688 | 93.24 | 95.34 | 97.70 | 92.41 |
| Pv39 | 7 639 474 | 7 613 382 | 1 145 921 100 | 1 090 946 618 | 7 174 002 | 94.23 | 95.20 | 97.83 | 92.78 |
| Pv40 | 7 361 614 | 7 337 070 | 1 104 242 100 | 1 066 060 554 | 6 703 574 | 91.37 | 96.54 | 97.57 | 91.86 |
| Pv41 | 7 505 308 | 7 480 256 | 1 125 796 200 | 1 080 668 216 | 6 893 819 | 92.16 | 95.99 | 97.72 | 92.44 |
| Pv42 | 7 348 356 | 7 324 392 | 1 102 253 400 | 1 063 679 164 | 6 768 694 | 92.41 | 96.50 | 97.30 | 91.03 |
| Pv43 | 6 587 980 | 6 565 632 | 988 197 000 | 946 190 986 | 6 004 408 | 91.45 | 95.75 | 97.54 | 91.90 |
| Pv44 | 7 403 192 | 7 379 070 | 1 110 478 800 | 1 058 345 682 | 6 901 485 | 93.53 | 95.31 | 97.57 | 91.97 |
| Pv45 | 7 764 072 | 7 737 444 | 1 164 610 800 | 1 108 132 120 | 7 056 881 | 91.20 | 95.15 | 97.79 | 92.62 |
| Pv46 | 4 830 616 | 4 814 580 | 724 592 400 | 696 981 244 | 4 397 219 | 91.33 | 96.19 | 97.88 | 93.05 |
| Pv47 | 7 547 974 | 7 522 116 | 1 132 196 100 | 1 078 148 092 | 7 025 146 | 93.39 | 95.23 | 97.88 | 92.86 |
| Pv48 | 6 978 052 | 6 954 442 | 1 046 707 800 | 1 001 538 926 | 6 352 621 | 91.35 | 95.68 | 97.79 | 92.65 |
| Pv49 | 6 826 836 | 6 804 008 | 1 024 025 400 | 985 507 780 | 6 214 244 | 91.33 | 96.24 | 97.76 | 92.53 |
| Pv50 | 5 268 676 | 5 251 028 | 790 301 400 | 754 087 146 | 4 785 144 | 91.13 | 95.42 | 97.75 | 92.40 |
| Pv51 | 6 257 576 | 6 236 592 | 938 636 400 | 894 014 262 | 5 688 008 | 91.20 | 95.25 | 97.81 | 92.69 |
| Pv52 | 6 214 442 | 6 193 152 | 932 166 300 | 895 366 264 | 5 662 541 | 91.43 | 96.05 | 97.83 | 92.81 |
| Pv53 | 7 123 026 | 7 099 578 | 1 068 453 900 | 1 025 234 062 | 6 483 231 | 91.32 | 95.95 | 97.67 | 92.23 |
| Pv54 | 2 996 190 | 2 985 946 | 449 428 500 | 430 855 692 | 2 736 528 | 91.65 | 95.87 | 98.09 | 93.66 |
| Pv55 | 3 334 436 | 3 323 366 | 500 165 400 | 482 901 568 | 3 040 272 | 91.48 | 96.55 | 97.29 | 91.07 |
| Pv56 | 5 981 750 | 5 961 578 | 897 262 500 | 864 283 148 | 5 454 974 | 91.50 | 96.32 | 97.51 | 91.69 |
| Pv57 | 6 743 544 | 6 720 248 | 1 011 531 600 | 966 479 926 | 6 292 623 | 93.64 | 95.55 | 97.99 | 93.32 |
| Pv58 | 6 757 314 | 6 734 582 | 1 013 597 100 | 967 411 196 | 6 149 238 | 91.31 | 95.44 | 97.66 | 92.26 |
| Pv59 | 6 644 366 | 6 622 624 | 996 654 900 | 952 675 344 | 6 039 136 | 91.19 | 95.59 | 97.71 | 92.41 |
| Pv60 | 3 308 738 | 3 297 088 | 496 310 700 | 473 938 530 | 3 006 470 | 91.19 | 95.49 | 98.13 | 93.75 |
| Pv61 | 7 164 518 | 7 140 300 | 1 074 677 700 | 1 028 969 692 | 6 606 890 | 92.53 | 95.75 | 97.82 | 92.79 |
| Pv62 | 6 544 594 | 6 522 146 | 981 689 100 | 937 859 972 | 5 945 974 | 91.17 | 95.54 | 97.57 | 91.96 |
| Pv63 | 6 262 790 | 6 242 050 | 939 418 500 | 901 268 214 | 5 721 647 | 91.66 | 95.94 | 97.61 | 92.12 |
| Pv64 | 7 756 110 | 7 729 588 | 1 163 416 500 | 1 110 586 330 | 7 062 279 | 91.37 | 95.46 | 97.69 | 92.31 |
| Pv65 | 7 212 746 | 7 189 088 | 1 081 911 900 | 1 038 137 070 | 6 569 530 | 91.38 | 95.95 | 97.64 | 92.24 |
| Pv66 | 6 842 464 | 6 819 456 | 1 026 369 600 | 980 771 280 | 6 227 660 | 91.32 | 95.56 | 97.80 | 92.66 |
| Pv67 | 5 491 250 | 5 472 356 | 823 687 500 | 786 889 766 | 5 007 569 | 91.51 | 95.53 | 98.22 | 94.03 |
| Pv68 | 8 598 260 | 8 569 182 | 1 289 739 000 | 1 225 043 812 | 7 824 318 | 91.31 | 94.98 | 97.96 | 93.23 |
| Pv69 | 7 098 136 | 7 074 072 | 1 064 720 400 | 1 021 245 826 | 6 476 276 | 91.55 | 95.92 | 97.89 | 92.98 |
| Pv70 | 6 744 486 | 6 721 580 | 1 011 672 900 | 970 160 098 | 6 153 450 | 91.55 | 95.90 | 97.50 | 91.86 |
| Pv71 | 6 954 908 | 6 932 014 | 1 043 236 200 | 997 598 058 | 6 333 065 | 91.36 | 95.63 | 97.56 | 91.86 |
| Pv72 | 6 947 998 | 6 924 592 | 1 042 199 700 | 995 918 234 | 6 390 351 | 92.28 | 95.56 | 97.92 | 93.06 |
| Pv73 | 11 385 340 | 11 365 232 | 1 707 801 000 | 1 521 977 790 | 10 427 776 | 91.75 | 89.12 | 98.37 | 94.62 |
| Pv74 | 6 704 026 | 6 681 088 | 1 005 603 900 | 966 719 208 | 6 098 771 | 91.28 | 96.13 | 97.64 | 92.13 |
| Pv75 | 15 259 508 | 15 229 100 | 2 288 926 200 | 2 115 314 516 | 13 656 417 | 89.67 | 92.42 | 98.24 | 94.14 |
| Pv76 | 6 308 324 | 6 287 108 | 946 248 600 | 904 804 666 | 5 822 194 | 92.61 | 95.62 | 98.04 | 93.56 |
| Pv77 | 5 934 858 | 5 915 132 | 890 228 700 | 860 418 210 | 5 605 331 | 94.76 | 96.65 | 97.57 | 91.79 |
| Pv78 | 7 053 330 | 7 029 220 | 1 057 999 500 | 1 014 776 334 | 6 427 694 | 91.44 | 95.91 | 97.83 | 92.76 |
| Pv79 | 6 626 144 | 6 603 632 | 993 921 600 | 954 116 282 | 6 234 280 | 94.41 | 96.00 | 97.74 | 92.42 |
| Pv80 | 7 477 584 | 7 452 982 | 1 121 637 600 | 1 073 606 700 | 6 968 107 | 93.49 | 95.72 | 97.59 | 92.08 |
| Pv81 | 6 965 426 | 6 941 310 | 1 044 813 900 | 1 002 425 192 | 6 527 168 | 94.03 | 95.94 | 98.23 | 94.06 |
| Pv82 | 7 585 280 | 7 559 696 | 1 137 792 000 | 1 094 479 824 | 6 903 582 | 91.32 | 96.19 | 97.73 | 92.43 |
| Pv83 | 8 660 626 | 8 630 762 | 1 299 093 900 | 1 241 602 578 | 7 908 789 | 91.63 | 95.57 | 97.98 | 93.28 |
| Pv84 | 7 290 040 | 7 264 834 | 1 093 506 000 | 1 048 919 858 | 6 648 774 | 91.52 | 95.92 | 97.60 | 92.08 |
| Pv85 | 7 969 190 | 7 941 526 | 1 195 378 500 | 1 149 028 462 | 7 385 800 | 93.00 | 96.12 | 97.99 | 93.34 |
| Pv86 | 7 584 448 | 7 558 542 | 1 137 667 200 | 1 082 746 918 | 6 911 071 | 91.43 | 95.17 | 98.27 | 94.17 |
| Pv87 | 7 584 774 | 7 558 902 | 1 137 716 100 | 1 083 325 126 | 6 884 039 | 91.07 | 95.22 | 97.91 | 93.08 |
| Pv88 | 7 352 392 | 7 327 872 | 1 102 858 800 | 1 066 357 808 | 6 738 930 | 91.96 | 96.69 | 97.38 | 91.41 |
| Pv89 | 6 766 290 | 6 743 796 | 1 014 943 500 | 967 162 154 | 6 191 315 | 91.81 | 95.29 | 97.74 | 92.52 |
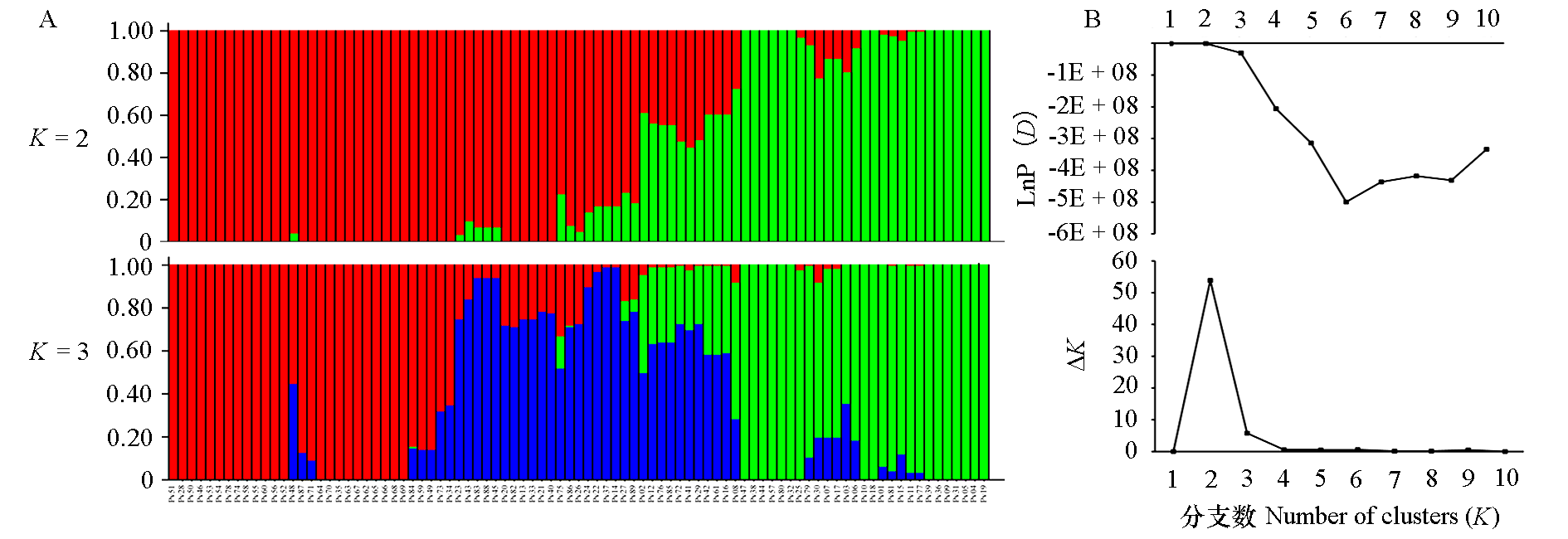
图4 菜豆89份品种的群体遗传结构分析 A:K = 2(最佳K值)和K = 3时的群体结构,每条竖线代表一个品种,不同颜色代表基因型来源不同;B:K = 2 ~ 10 的平均后验概率[LnP(D)]值和相对应的ΔK。
Fig. 4 Population structure analysis of the 89 common bean cultivars A:Histogram of the STRUCTURE analysis for the model with K = 2(showing the highest ΔK)and K = 3. A vertical bar represents a single individual and each color corresponds to a suggested cluster. B:Plots of the mean posterior probability[LnP(D)]values of each K and the corresponding ΔK statistics.
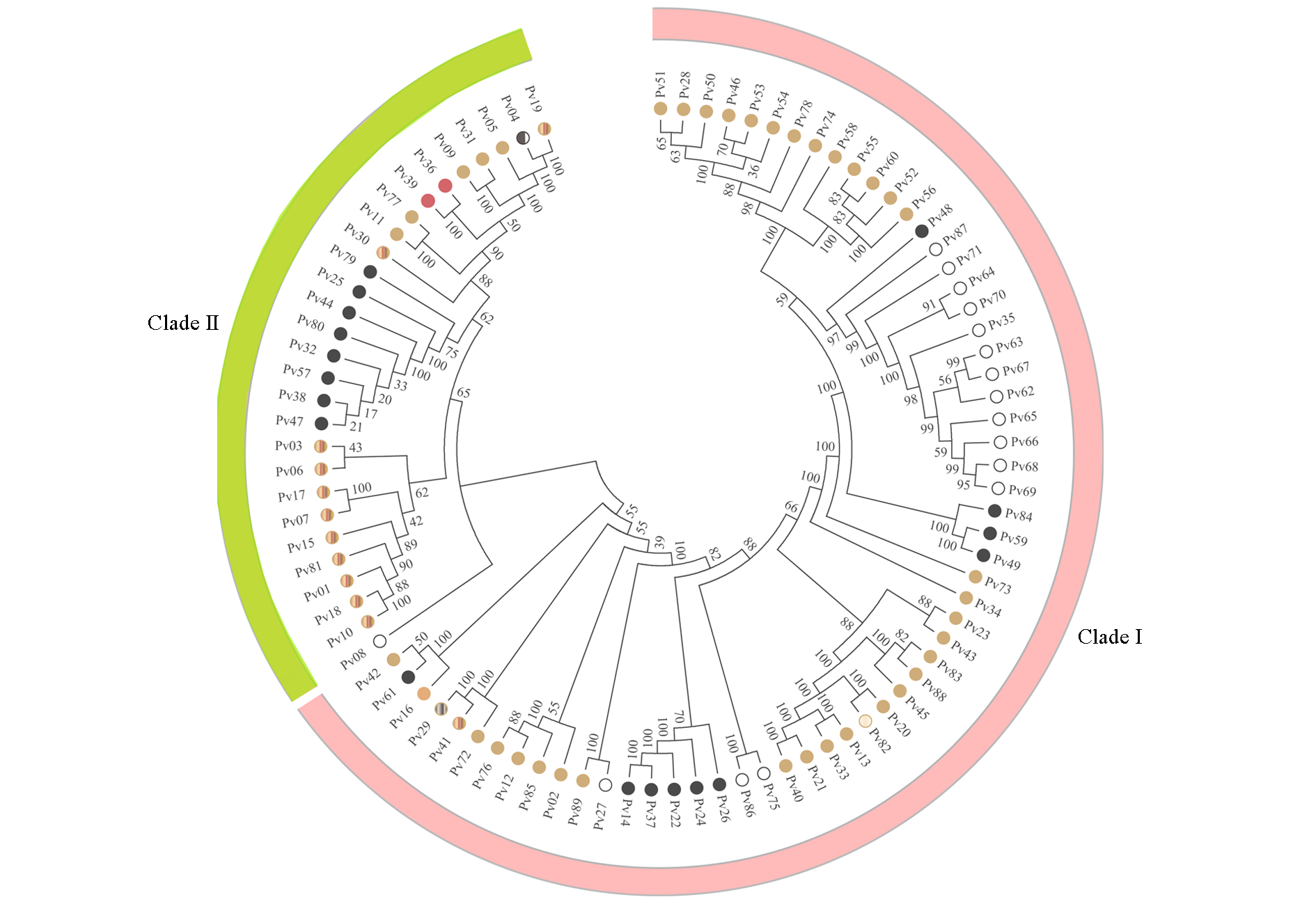
图5 基于10K液相芯片技术的89个菜豆品种聚类图 每个个体对应圆圈的颜色表示种皮的颜色。
Fig. 5 Cluster analysis diagram of 89 common cultivars based on 10K liquid chip The colour of the circles indicates the colour of the seed coat.
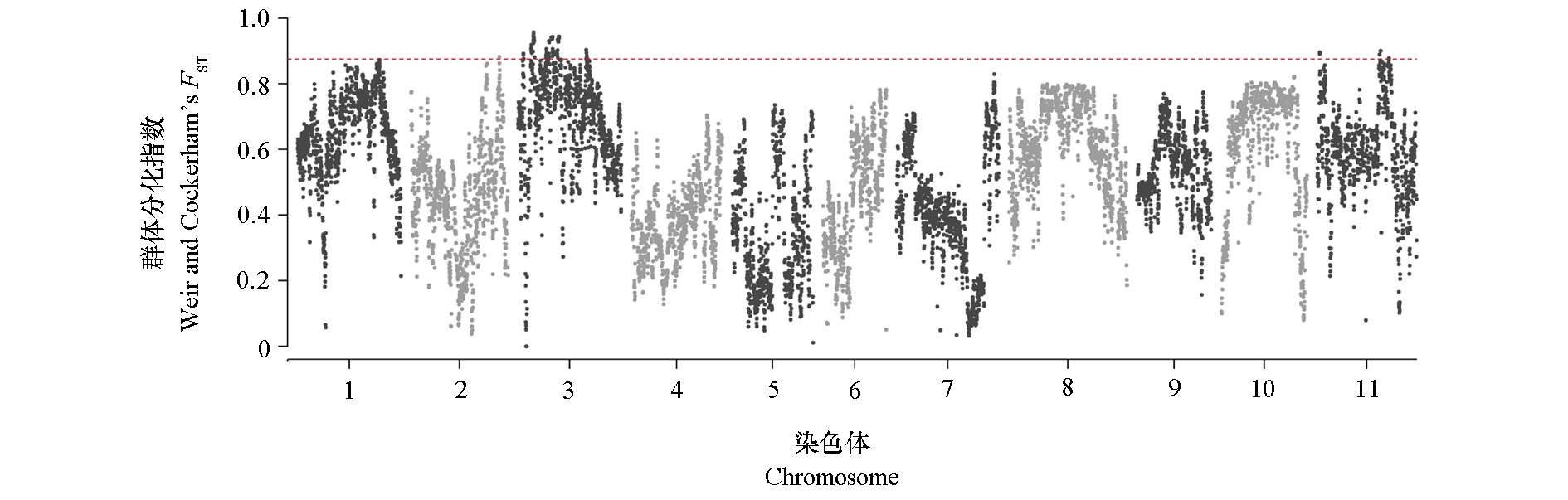
图6 菜豆2个品种类群之间的选择位点扫描 红色虚线代表的是1%的显著性阈值。
Fig. 6 Selective-sweep signals among the two common bean groups The red dashed lines represent the thresholds(top 1% of FST values).
| 选择信号 Selective signal | 染色体 Chr. | 开始位置 Start position | 终止位置 End position | 群体分化 指数 FST | 选择信号 Selective signal | 染色体 Chr. | 开始位置 Start position | 终止位置 End position | 群体分化 指数 FST |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 44 650 001 | 45 150 000 | 0.722 | 22 | 3 | 7 900 001 | 8 400 000 | 0.752 |
| 2 | 2 | 44 700 001 | 45 200 000 | 0.722 | 23 | 3 | 7 950 001 | 8 450 000 | 0.741 |
| 3 | 3 | 2 600 001 | 3 100 000 | 0.669 | 24 | 3 | 8 000 001 | 8 500 000 | 0.761 |
| 4 | 3 | 2 650 001 | 3 150 000 | 0.667 | 25 | 3 | 8 050 001 | 8 550 000 | 0.761 |
| 5 | 3 | 2 700 001 | 3 200 000 | 0.753 | 26 | 3 | 8 100 001 | 8 600 000 | 0.681 |
| 6 | 3 | 6 550 001 | 7 050 000 | 0.880 | 27 | 3 | 8 150 001 | 8 650 000 | 0.679 |
| 7 | 3 | 6 600 001 | 7 100 000 | 0.877 | 28 | 3 | 8 200 001 | 8 700 000 | 0.633 |
| 8 | 3 | 6 650 001 | 7 150 000 | 0.904 | 29 | 3 | 14 100 001 | 14 600 000 | 0.769 |
| 9 | 3 | 6 700 001 | 7 200 000 | 0.830 | 30 | 3 | 14 150 001 | 14 650 000 | 0.784 |
| 10 | 3 | 6 750 001 | 7 250 000 | 0.830 | 31 | 3 | 14 200 001 | 14 700 000 | 0.821 |
| 11 | 3 | 6 800 001 | 7 300 000 | 0.827 | 32 | 3 | 14 250 001 | 14 750 000 | 0.810 |
| 12 | 3 | 6 850 001 | 7 350 000 | 0.827 | 33 | 3 | 14 300 001 | 14 800 000 | 0.726 |
| 13 | 3 | 7 450 001 | 7 950 000 | 0.565 | 34 | 3 | 14 350 001 | 14 850 000 | 0.725 |
| 14 | 3 | 7 500 001 | 8 000 000 | 0.565 | 35 | 3 | 14 400 001 | 14 900 000 | 0.725 |
| 15 | 3 | 7 550 001 | 8 050 000 | 0.560 | 36 | 3 | 14 450 001 | 14 950 000 | 0.786 |
| 16 | 3 | 7 600 001 | 8 100 000 | 0.657 | 37 | 3 | 15 400 001 | 15 900 000 | 0.723 |
| 17 | 3 | 7 650 001 | 8 150 000 | 0.668 | 38 | 3 | 15 450 001 | 15 950 000 | 0.632 |
| 18 | 3 | 7 700 001 | 8 200 000 | 0.668 | 39 | 3 | 15 500 001 | 16 000 000 | 0.714 |
| 19 | 3 | 7 750 001 | 8 250 000 | 0.763 | 40 | 3 | 15 550 001 | 16 050 000 | 0.769 |
| 20 | 3 | 7 800 001 | 8 300 000 | 0.862 | 41 | 3 | 15 600 001 | 16 100 000 | 0.726 |
| 21 | 3 | 7 850 001 | 8 350 000 | 0.850 | 42 | 3 | 15 650 001 | 16 150 000 | 0.730 |
| 43 | 3 | 15 700 001 | 16 200 000 | 0.671 | 74 | 3 | 20 200 001 | 20 700 000 | 0.798 |
| 44 | 3 | 16 800 001 | 17 300 000 | 0.941 | 75 | 3 | 20 250 001 | 20 750 000 | 0.768 |
| 45 | 3 | 16 850 001 | 17 350 000 | 0.941 | 76 | 3 | 20 300 001 | 20 800 000 | 0.729 |
| 46 | 3 | 16 900 001 | 17 400 000 | 0.940 | 77 | 3 | 20 350 001 | 20 850 000 | 0.729 |
| 47 | 3 | 16 950 001 | 17 450 000 | 0.940 | 78 | 3 | 20 400 001 | 20 900 000 | 0.942 |
| 48 | 3 | 17 000 001 | 17 500 000 | 0.940 | 79 | 3 | 20 450 001 | 20 950 000 | 0.943 |
| 49 | 3 | 17 050 001 | 17 550 000 | 0.940 | 80 | 3 | 20 500 001 | 21 000 000 | 0.942 |
| 50 | 3 | 17 100 001 | 17 600 000 | 0.936 | 81 | 3 | 20 550 001 | 21 050 000 | 0.944 |
| 51 | 3 | 17 150 001 | 17 650 000 | 0.937 | 82 | 3 | 20 600 001 | 21 100 000 | 0.944 |
| 52 | 3 | 17 200 001 | 17 700 000 | 0.937 | 83 | 3 | 20 650 001 | 21 150 000 | 0.944 |
| 53 | 3 | 17 250 001 | 17 750 000 | 0.940 | 84 | 3 | 20 700 001 | 21 200 000 | 0.943 |
| 54 | 3 | 17 300 001 | 17 800 000 | 0.943 | 85 | 3 | 20 750 001 | 21 250 000 | 0.940 |
| 55 | 3 | 17 350 001 | 17 850 000 | 0.790 | 86 | 3 | 34 300 001 | 34 800 000 | 0.703 |
| 56 | 3 | 17 400 001 | 17 900 000 | 0.657 | 87 | 3 | 34 350 001 | 34 850 000 | 0.703 |
| 57 | 3 | 17 450 001 | 17 950 000 | 0.699 | 88 | 3 | 34 400 001 | 34 900 000 | 0.750 |
| 58 | 3 | 17 500 001 | 18 000 000 | 0.699 | 89 | 3 | 34 450 001 | 34 950 000 | 0.749 |
| 59 | 3 | 17 550 001 | 18 050 000 | 0.699 | 90 | 3 | 34 500 001 | 35 000 000 | 0.728 |
| 60 | 3 | 17 600 001 | 18 100 000 | 0.701 | 91 | 3 | 34 650 001 | 35 150 000 | 0.653 |
| 61 | 3 | 17 650 001 | 18 150 000 | 0.661 | 92 | 3 | 34 700 001 | 35 200 000 | 0.654 |
| 62 | 3 | 17 700 001 | 18 200 000 | 0.661 | 93 | 3 | 34 800 001 | 35 300 000 | 0.758 |
| 63 | 3 | 17 900 001 | 18 400 000 | 0.762 | 94 | 11 | 1 300 001 | 1 800 000 | 0.824 |
| 64 | 3 | 18 750 001 | 19 250 000 | 0.777 | 95 | 11 | 1 350 001 | 1 850 000 | 0.838 |
| 65 | 3 | 18 800 001 | 19 300 000 | 0.797 | 96 | 11 | 1 400 001 | 1 900 000 | 0.829 |
| 66 | 3 | 19 800 001 | 20 300 000 | 0.815 | 97 | 11 | 1 450 001 | 1 950 000 | 0.832 |
| 67 | 3 | 19 850 001 | 20 350 000 | 0.798 | 98 | 11 | 31 450 001 | 31 950 000 | 0.888 |
| 68 | 3 | 19 900 001 | 20 400 000 | 0.747 | 99 | 11 | 32 000 001 | 32 500 000 | 0.900 |
| 69 | 3 | 19 950 001 | 20 450 000 | 0.819 | 100 | 11 | 32 050 001 | 32 550 000 | 0.899 |
| 70 | 3 | 20 000 001 | 20 500 000 | 0.819 | 101 | 11 | 32 100 001 | 32 600 000 | 0.899 |
| 71 | 3 | 20 050 001 | 20 550 000 | 0.834 | 102 | 11 | 36 200 001 | 36 700 000 | 0.764 |
| 72 | 3 | 20 100 001 | 20 600 000 | 0.819 | 103 | 11 | 36 300 001 | 36 800 000 | 0.780 |
| 73 | 3 | 20 150 001 | 20 650 000 | 0.819 |
表3 103个处于强烈选择状态的区域信息
Table 3 Information of 103 regions under strong selection
| 选择信号 Selective signal | 染色体 Chr. | 开始位置 Start position | 终止位置 End position | 群体分化 指数 FST | 选择信号 Selective signal | 染色体 Chr. | 开始位置 Start position | 终止位置 End position | 群体分化 指数 FST |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 44 650 001 | 45 150 000 | 0.722 | 22 | 3 | 7 900 001 | 8 400 000 | 0.752 |
| 2 | 2 | 44 700 001 | 45 200 000 | 0.722 | 23 | 3 | 7 950 001 | 8 450 000 | 0.741 |
| 3 | 3 | 2 600 001 | 3 100 000 | 0.669 | 24 | 3 | 8 000 001 | 8 500 000 | 0.761 |
| 4 | 3 | 2 650 001 | 3 150 000 | 0.667 | 25 | 3 | 8 050 001 | 8 550 000 | 0.761 |
| 5 | 3 | 2 700 001 | 3 200 000 | 0.753 | 26 | 3 | 8 100 001 | 8 600 000 | 0.681 |
| 6 | 3 | 6 550 001 | 7 050 000 | 0.880 | 27 | 3 | 8 150 001 | 8 650 000 | 0.679 |
| 7 | 3 | 6 600 001 | 7 100 000 | 0.877 | 28 | 3 | 8 200 001 | 8 700 000 | 0.633 |
| 8 | 3 | 6 650 001 | 7 150 000 | 0.904 | 29 | 3 | 14 100 001 | 14 600 000 | 0.769 |
| 9 | 3 | 6 700 001 | 7 200 000 | 0.830 | 30 | 3 | 14 150 001 | 14 650 000 | 0.784 |
| 10 | 3 | 6 750 001 | 7 250 000 | 0.830 | 31 | 3 | 14 200 001 | 14 700 000 | 0.821 |
| 11 | 3 | 6 800 001 | 7 300 000 | 0.827 | 32 | 3 | 14 250 001 | 14 750 000 | 0.810 |
| 12 | 3 | 6 850 001 | 7 350 000 | 0.827 | 33 | 3 | 14 300 001 | 14 800 000 | 0.726 |
| 13 | 3 | 7 450 001 | 7 950 000 | 0.565 | 34 | 3 | 14 350 001 | 14 850 000 | 0.725 |
| 14 | 3 | 7 500 001 | 8 000 000 | 0.565 | 35 | 3 | 14 400 001 | 14 900 000 | 0.725 |
| 15 | 3 | 7 550 001 | 8 050 000 | 0.560 | 36 | 3 | 14 450 001 | 14 950 000 | 0.786 |
| 16 | 3 | 7 600 001 | 8 100 000 | 0.657 | 37 | 3 | 15 400 001 | 15 900 000 | 0.723 |
| 17 | 3 | 7 650 001 | 8 150 000 | 0.668 | 38 | 3 | 15 450 001 | 15 950 000 | 0.632 |
| 18 | 3 | 7 700 001 | 8 200 000 | 0.668 | 39 | 3 | 15 500 001 | 16 000 000 | 0.714 |
| 19 | 3 | 7 750 001 | 8 250 000 | 0.763 | 40 | 3 | 15 550 001 | 16 050 000 | 0.769 |
| 20 | 3 | 7 800 001 | 8 300 000 | 0.862 | 41 | 3 | 15 600 001 | 16 100 000 | 0.726 |
| 21 | 3 | 7 850 001 | 8 350 000 | 0.850 | 42 | 3 | 15 650 001 | 16 150 000 | 0.730 |
| 43 | 3 | 15 700 001 | 16 200 000 | 0.671 | 74 | 3 | 20 200 001 | 20 700 000 | 0.798 |
| 44 | 3 | 16 800 001 | 17 300 000 | 0.941 | 75 | 3 | 20 250 001 | 20 750 000 | 0.768 |
| 45 | 3 | 16 850 001 | 17 350 000 | 0.941 | 76 | 3 | 20 300 001 | 20 800 000 | 0.729 |
| 46 | 3 | 16 900 001 | 17 400 000 | 0.940 | 77 | 3 | 20 350 001 | 20 850 000 | 0.729 |
| 47 | 3 | 16 950 001 | 17 450 000 | 0.940 | 78 | 3 | 20 400 001 | 20 900 000 | 0.942 |
| 48 | 3 | 17 000 001 | 17 500 000 | 0.940 | 79 | 3 | 20 450 001 | 20 950 000 | 0.943 |
| 49 | 3 | 17 050 001 | 17 550 000 | 0.940 | 80 | 3 | 20 500 001 | 21 000 000 | 0.942 |
| 50 | 3 | 17 100 001 | 17 600 000 | 0.936 | 81 | 3 | 20 550 001 | 21 050 000 | 0.944 |
| 51 | 3 | 17 150 001 | 17 650 000 | 0.937 | 82 | 3 | 20 600 001 | 21 100 000 | 0.944 |
| 52 | 3 | 17 200 001 | 17 700 000 | 0.937 | 83 | 3 | 20 650 001 | 21 150 000 | 0.944 |
| 53 | 3 | 17 250 001 | 17 750 000 | 0.940 | 84 | 3 | 20 700 001 | 21 200 000 | 0.943 |
| 54 | 3 | 17 300 001 | 17 800 000 | 0.943 | 85 | 3 | 20 750 001 | 21 250 000 | 0.940 |
| 55 | 3 | 17 350 001 | 17 850 000 | 0.790 | 86 | 3 | 34 300 001 | 34 800 000 | 0.703 |
| 56 | 3 | 17 400 001 | 17 900 000 | 0.657 | 87 | 3 | 34 350 001 | 34 850 000 | 0.703 |
| 57 | 3 | 17 450 001 | 17 950 000 | 0.699 | 88 | 3 | 34 400 001 | 34 900 000 | 0.750 |
| 58 | 3 | 17 500 001 | 18 000 000 | 0.699 | 89 | 3 | 34 450 001 | 34 950 000 | 0.749 |
| 59 | 3 | 17 550 001 | 18 050 000 | 0.699 | 90 | 3 | 34 500 001 | 35 000 000 | 0.728 |
| 60 | 3 | 17 600 001 | 18 100 000 | 0.701 | 91 | 3 | 34 650 001 | 35 150 000 | 0.653 |
| 61 | 3 | 17 650 001 | 18 150 000 | 0.661 | 92 | 3 | 34 700 001 | 35 200 000 | 0.654 |
| 62 | 3 | 17 700 001 | 18 200 000 | 0.661 | 93 | 3 | 34 800 001 | 35 300 000 | 0.758 |
| 63 | 3 | 17 900 001 | 18 400 000 | 0.762 | 94 | 11 | 1 300 001 | 1 800 000 | 0.824 |
| 64 | 3 | 18 750 001 | 19 250 000 | 0.777 | 95 | 11 | 1 350 001 | 1 850 000 | 0.838 |
| 65 | 3 | 18 800 001 | 19 300 000 | 0.797 | 96 | 11 | 1 400 001 | 1 900 000 | 0.829 |
| 66 | 3 | 19 800 001 | 20 300 000 | 0.815 | 97 | 11 | 1 450 001 | 1 950 000 | 0.832 |
| 67 | 3 | 19 850 001 | 20 350 000 | 0.798 | 98 | 11 | 31 450 001 | 31 950 000 | 0.888 |
| 68 | 3 | 19 900 001 | 20 400 000 | 0.747 | 99 | 11 | 32 000 001 | 32 500 000 | 0.900 |
| 69 | 3 | 19 950 001 | 20 450 000 | 0.819 | 100 | 11 | 32 050 001 | 32 550 000 | 0.899 |
| 70 | 3 | 20 000 001 | 20 500 000 | 0.819 | 101 | 11 | 32 100 001 | 32 600 000 | 0.899 |
| 71 | 3 | 20 050 001 | 20 550 000 | 0.834 | 102 | 11 | 36 200 001 | 36 700 000 | 0.764 |
| 72 | 3 | 20 100 001 | 20 600 000 | 0.819 | 103 | 11 | 36 300 001 | 36 800 000 | 0.780 |
| 73 | 3 | 20 150 001 | 20 650 000 | 0.819 |
| [1] |
doi: 10.3389/fpls.2017.00722 pmid: 28533789 |
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
doi: 10.1111/j.1365-294X.2005.02553.x pmid: 15969739 |
| [7] |
|
|
郭永田. 2014. 中国食用豆产业的经济分析[博士论文]. 武汉: 华中农业大学.
|
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
doi: 10.16420/j.issn.0513-353x.2022-0981 |
|
刘承浪, 冯迪, 曹宗洪, 闫素云, 张永青, 徐祥增, 高世德, 叶俊丽, 柴利军, 谢宗周, 邓秀新. 2023. 东试早柚与沙田柚和水晶柚遗传背景比较. 园艺学报, 50 (11):2337-2349.
doi: 10.16420/j.issn.0513-353x.2022-0981 |
|
| [15] |
|
|
刘大亮. 2020. 9种豆科植物含env结构的LTR反转座元件的注释及菜豆RBIP分子标记开发[博士论文]. 南京: 南京农业大学.
|
|
| [16] |
|
| [17] |
|
|
栾非时. 2000. 菜豆种质资源遗传多样性的研究[博士论文]. 哈尔滨: 东北农业大学.
|
|
| [18] |
|
| [19] |
Mahiya-Farooq,
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
|
彭丽娟, 牟柯澴, 张健伟, 赵兆, 山诗瑶, 薛新如, 朱丽, 许琳玉, 李焕秀. 2022. 基于SSR及ISSR标记的菜豆遗传多样性分析. 分子植物育种, 20 (21):7161-7173.
|
|
| [24] |
doi: 10.1093/genetics/155.2.945 pmid: 10835412 |
| [25] |
|
| [26] |
doi: S1674-2052(17)30174-0 pmid: 28669791 |
| [27] |
doi: 10.16420/j.issn.0513-353x.2021-1046 |
|
任海龙, 许东林, 张晶, 邹集文, 李光光, 周贤玉, 肖婉钰, 孙艺嘉. 2023. 菜薹KASP-SNP指纹图谱构建及品种鉴定. 园艺学报, 50 (2):307-318.
doi: 10.16420/j.issn.0513-353x.2021-1046 |
|
| [28] |
doi: 10.1038/ng.3008 pmid: 24908249 |
| [29] |
|
| [30] |
|
| [31] |
doi: 10.1186/s13059-016-0883-6 pmid: 26911872 |
| [32] |
doi: 10.13430/j.cnki.jpgr.20200309002 |
|
王富强, 樊秀彩, 张颖, 刘崇怀, 姜建福. 2020. SNP 分子标记在作物品种鉴定中的应用和展望. 植物遗传资源学报, 21 (5):1308-1320.
doi: 10.13430/j.cnki.jpgr.20200309002 |
|
| [33] |
|
|
王述民, 段醒男, 丁国庆, 王晓鸣. 1999. 普通菜豆种质资源的收集与评价. 作物品种资源,(3):52-53.
|
|
| [34] |
|
|
汪宝根, 董君暘, 汪颖, 李素娟, 王尖, 鲁忠富, 吴晓花, 李国景, 吴新义. 2022. 浙江省地方菜豆种质资源鉴定与遗传多样性分析. 浙江农业学报, 34 (11):2416-2427.
doi: 10.3969/j.issn.1004-1524.2022.11.11 |
|
| [35] |
doi: 10.1038/s41588-019-0546-0 pmid: 31873299 |
| [36] |
|
|
武晶. 2021. 普通菜豆基因组研究进展. 四川农业大学学报, 39 (1):4-10.
|
|
| [37] |
|
|
肖靖, 李斌, 石晓华, 鄂成林, 管洪波, 凤桐. 2016. 普通菜豆种质资源研究现状及进展. 北方园艺, 40 (15):194-198.
|
|
| [38] |
|
|
徐云碧, 杨泉女, 郑洪建, 许彦芬, 桑志勤, 郭子锋, 彭海, 张丛, 蓝昊发, 王蕴波, 吴坤生, 陶家军, 张嘉楠. 2020. 靶向测序基因型检测(GBTS)技术及其应用. 中国农业科学, 53 (15):2983-3004.
doi: 10.3864/j.issn.0578-1752.2020.15.001 |
|
| [39] |
|
| [40] |
|
|
张赤红, 王述民. 2005. 利用SSR标记评价普通菜豆种质遗传多样性. 作物学报, 31 (5):619-627.
|
|
| [41] |
|
|
张赤红, 王述民, 曹永生, 宗绪晓. 2005. 普通菜豆种质资源形态多样性鉴定与分类研究. 中国农业科学, 38 (1):27-32.
|
|
| [42] |
|
| [43] |
doi: 10.3864/j.issn.0578-1752.2012.03.001 |
|
张立娜, 曹桂兰, 韩龙植. 2012. 利用SSR标记揭示中国粳稻地方品种遗传多样性. 中国农业科学, 45 (3):405-413.
doi: 10.3864/j.issn.0578-1752.2012.03.001 |
|
| [44] |
|
|
郑卓杰. 1997. 中国食用豆类学. 北京: 中国农业出版社.
|
| [1] | 秦子璐, 徐正康, 戴晓港, 陈赢男. 望春玉兰种质资源遗传多样性分析与核心种质构建[J]. 园艺学报, 2024, 51(8): 1823-1832. |
| [2] | 游 平, 杨 进, 周 俊, 黄爱军, 鲍敏丽, 易 龙, . 柑橘黄龙病菌原噬菌体的遗传多样性分析[J]. 园艺学报, 2024, 51(4): 727-736. |
| [3] | 尤园园, 王帅, 方莹莹, 齐诚, 李淑培, 张映, 陈钰辉, 刘伟, 刘富中, 舒金帅. 茄子全基因组InDel变异特征及分子标记开发和应用[J]. 园艺学报, 2024, 51(3): 520-532. |
| [4] | 孙挺, 蒋茹佳, 施政, Menachem Moshelion, 孙玉东, 程瑞, 徐沛. 菜豆“水分利用效率黄金时段”量化解析方法的建立及应用[J]. 园艺学报, 2024, 51(3): 656-668. |
| [5] | 罗新锐, 张晓旭, 王玉萍, 王 智, 马媛媛, 周丙月. 普通菜豆Trihelix基因家族鉴定及表达分析[J]. 园艺学报, 2024, 51(12): 2775-2790. |
| [6] | 陈方策, 胡平正, 解为玮, 王钲彭, 钟创南, 李海炎, 何业华, 彭泽, 万保雄, 刘朝阳. 基于重测序的中国李基因组InDel标记的开发及应用[J]. 园艺学报, 2024, 51(11): 2510-2522. |
| [7] | 王荣波, 王海云, 张前荣, 蔡松龄, 李本金, 张美祥, 刘裴清. 福建省番茄青枯病菌遗传多样性分析及其抗性砧木的筛选[J]. 园艺学报, 2024, 51(11): 2540-2554. |
| [8] | 李春牛, 苏 群, 李先民, 黄展文, 孙明艳, 卢家仕, 王虹妍, 卜朝阳. 茉莉花全基因组SSR标记开发及其亲缘关系鉴定[J]. 园艺学报, 2024, 51(10): 2343-2357. |
| [9] | 关夏玉, 陈细红, 郭菁, 吕浩阳, 梁晨媛, 高芳銮. 建兰花叶病毒的遗传变异及其寄主适应性[J]. 园艺学报, 2024, 51(1): 203-212. |
| [10] | 王瑞, 洪文娟, 罗华, 赵丽娜, 陈颖, 王君. 石榴品种SSR指纹图谱构建及杂种父本鉴定[J]. 园艺学报, 2023, 50(2): 265-278. |
| [11] | 王梦梦, 孙德岭, 陈锐, 杨迎霞, 张冠, 吕明杰, 王倩, 谢添羽, 牛国保, 单晓政, 谭津, 姚星伟. 花椰菜核心种质的构建与评价[J]. 园艺学报, 2023, 50(2): 421-431. |
| [12] | 段开行, 王晓玲, 毛永民, 王瑶, 任勇响, 任柳柳, 申连英. 酸枣种质资源果实数量性状遗传多样性分析[J]. 园艺学报, 2023, 50(12): 2568-2576. |
| [13] | 谢倩, 张诗艳, 江来, 丁明月, 刘玲玲, 吴如健, 陈清西. 橄榄转录组SSR信息分析及分子标记开发与应用[J]. 园艺学报, 2023, 50(11): 2350-2364. |
| [14] | 苏群, 王虹妍, 刘俊, 李春牛, 卜朝阳, 林玉玲, 卢家仕, 赖钟雄. 基于SSR荧光标记构建睡莲核心种质[J]. 园艺学报, 2023, 50(10): 2128-2138. |
| [15] | 刘志远, 徐兆生, 张合龙, 钱 伟. 无筋菜豆新品种‘蔬豆6号’[J]. 园艺学报, 2022, 49(S2): 135-136. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司