
园艺学报 ›› 2022, Vol. 49 ›› Issue (8): 1772-1784.doi: 10.16420/j.issn.0513-353x.2021-0389
蒋思思1, 袁军1, 周文君1, 钮根花2, 周俊琴1,*( )
)
收稿日期:2021-12-21
修回日期:2022-03-08
出版日期:2022-08-25
发布日期:2022-09-05
通讯作者:
周俊琴
E-mail:zhoujunqin@csuft.edu.cn
基金资助:
JIANG Sisi1, YUAN Jun1, ZHOU Wenjun1, NIU Genhua2, ZHOU Junqin1,*( )
)
Received:2021-12-21
Revised:2022-03-08
Online:2022-08-25
Published:2022-09-05
Contact:
ZHOU Junqin
E-mail:zhoujunqin@csuft.edu.cn
摘要:
利用Illumina平台对薄壳山核桃(Carya illinoinensis)品种‘金华’‘波尼’和1株实生古树单株的叶绿体全基因组进行测序,并对其特征进行分析。3个薄壳山核桃叶绿体基因组均呈典型的四分体结构,大小分别为160 819、162 611和160 762 bp,基因序列高度保守。三者均注释得到124个基因,包括79种蛋白编码基因、38个tRNA和7个rRNA基因;三者有相似的重复序列类型,在其叶绿体基因组序列中分别检测出74、74和73个SSR位点;系统发育分析表明,三者均与喙核桃(Annamocarya sinensis)亲缘关系最为密切。
中图分类号:
蒋思思, 袁军, 周文君, 钮根花, 周俊琴. 薄壳山核桃(Carya illinoinensis)叶绿体基因组及其特征分析[J]. 园艺学报, 2022, 49(8): 1772-1784.
JIANG Sisi, YUAN Jun, ZHOU Wenjun, NIU Genhua, ZHOU Junqin. Complete Chloroplast Genome Sequence and Characteristics Analysis of Carya illinoinensis[J]. Acta Horticulturae Sinica, 2022, 49(8): 1772-1784.
| 品种 Cultivar | LSC | SSC | IR | Genome | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 长度/bp Length | G + C/% | 长度/% Length | 长度/bp Length | G + C/% | 长度/% Length | 长度/bp Length | G + C/% | 长度/% Length | 长度/bp Length | |
| 金华 Jinhua | 90 022 | 33.74 | 55.98 | 18 791 | 29.89 | 11.68 | 26 003 | 42.58 | 16.17 | 160 819 |
| 波尼 Pawnee | 90 022 | 33.74 | 55.36 | 16 999 | 29.93 | 10.45 | 27 795 | 41.74 | 17.09 | 162 611 |
| 实生单株 Seedling individual | 89 964 | 33.75 | 55.96 | 18 792 | 29.88 | 11.69 | 26 003 | 42.58 | 16.17 | 160 762 |
表1 薄壳山核桃‘波尼’‘金华’和实生单株的叶绿体基因组特征综述
Table 1 Summary of the chloroplast genomes features of Carya illinoinensis‘Pawnee’‘Jinhua’and seedling individual
| 品种 Cultivar | LSC | SSC | IR | Genome | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 长度/bp Length | G + C/% | 长度/% Length | 长度/bp Length | G + C/% | 长度/% Length | 长度/bp Length | G + C/% | 长度/% Length | 长度/bp Length | |
| 金华 Jinhua | 90 022 | 33.74 | 55.98 | 18 791 | 29.89 | 11.68 | 26 003 | 42.58 | 16.17 | 160 819 |
| 波尼 Pawnee | 90 022 | 33.74 | 55.36 | 16 999 | 29.93 | 10.45 | 27 795 | 41.74 | 17.09 | 162 611 |
| 实生单株 Seedling individual | 89 964 | 33.75 | 55.96 | 18 792 | 29.88 | 11.69 | 26 003 | 42.58 | 16.17 | 160 762 |

图2 薄壳山核桃实生单株叶绿体全基因组图谱 内侧基因顺时针,外侧基因逆时针(Lohse et al.,2013)。
Fig. 2 Gene map of the complete chloroplast genome of Carya illinoinensis seedling individual Genes drawn inside the circle are transcribed clockwise,and those outsides are counter clockwise(Lohse et al.,2013).
| 基因分类 Category for gene | 基因分组 Group of gene | 基因名称 Name of gene |
|---|---|---|
| 自我复制 Self replication | rRNA | rrn16S,rrn23,rrn23S,rrn4.5,rrn4.5S,rrn5,rrn5S |
| tRNA | trnA-UGC*,trnC-GCA,trnD-GUC,trnE-UUC,trnF-GAA,trnfM-CAU,trnG-GCC*,trnG-UCC,trnH-GUG,trnI-CAU,trnI-GAU*,trnK-UUU*,trnL-CAA,trnL-UAA*,trnL-UAG,trnM-CAU,trnN-GUU,trnP-GGG,trnP-UGG,trnQ-UUG,trnR-ACG,trnR-UCU,trnS-CGA,trnS-GCU,trnS-GGA,trnStop-UUA,trnS-UGA,trnT-AAG,trnT-AGU,trnT-CAG,trnT-GGU,trnT-UAG,trnT-UGU,trnV-AAC,trnV-GAC,trnV-UAC*,trnW-CCA,trnY-GUA | |
| RNA聚合酶亚基 RNA polymerase | rpoA,rpoB,rpoC1*,rpoC2 | |
| 核糖体小亚基 Ribosomal protein(SSU) | rps2,rps3,rps4,rps7,rps8,rps11,rps12T,*,rps14,rps15,rps16*,rps18,rps19 | |
| 核糖体大亚基 Ribosomal protein(LSU) | rpl2*,rpl14,rpl16*,rpl20,rpl22,rpl23,rpl32,rpl33,rpl36 | |
| 光合作用相关 Genes for photo- synthesis | ATP 合酶 ATP synthase | atpA,atpB,atpE,atpF*,atpH,atpI |
| 光和系统Ⅰ Photosystem Ⅰ | psaA,psaB,psaC,psaI,psaJ | |
| 光和系统Ⅱ Photosystem Ⅱ | psbA,psbB,psbC,psbD,psbE,psbF,psbI,psbJ,psbK,psbH,psbL,psbM,psbN,psbT,psbZ,ycf3** | |
| 二磷酸核酮糖羧化酶亚基 Subunit of rubisco | rbcL | |
| 细胞色素复合物 Cytochrome b/f complex | petA,petB*,petD*,petG,petL,petN | |
| NADH脱氢酶 NADH-dehydrogenase | ndhA*,ndhB*,ndhC,ndhD,ndhE,ndhF,ndhG,ndhH,ndhI,ndhJ,ndhK | |
| 其他与未知功能 Other and unknown function | accD,ccsA,cemA,clpP**,matK,ycf1,ycf2,ycf4,ycf15a |
表2 薄壳山核桃叶绿体基因组注释基因列表
Table 2 List of genes annotated in the chloroplast genome of Carya illinoinensis sequenced
| 基因分类 Category for gene | 基因分组 Group of gene | 基因名称 Name of gene |
|---|---|---|
| 自我复制 Self replication | rRNA | rrn16S,rrn23,rrn23S,rrn4.5,rrn4.5S,rrn5,rrn5S |
| tRNA | trnA-UGC*,trnC-GCA,trnD-GUC,trnE-UUC,trnF-GAA,trnfM-CAU,trnG-GCC*,trnG-UCC,trnH-GUG,trnI-CAU,trnI-GAU*,trnK-UUU*,trnL-CAA,trnL-UAA*,trnL-UAG,trnM-CAU,trnN-GUU,trnP-GGG,trnP-UGG,trnQ-UUG,trnR-ACG,trnR-UCU,trnS-CGA,trnS-GCU,trnS-GGA,trnStop-UUA,trnS-UGA,trnT-AAG,trnT-AGU,trnT-CAG,trnT-GGU,trnT-UAG,trnT-UGU,trnV-AAC,trnV-GAC,trnV-UAC*,trnW-CCA,trnY-GUA | |
| RNA聚合酶亚基 RNA polymerase | rpoA,rpoB,rpoC1*,rpoC2 | |
| 核糖体小亚基 Ribosomal protein(SSU) | rps2,rps3,rps4,rps7,rps8,rps11,rps12T,*,rps14,rps15,rps16*,rps18,rps19 | |
| 核糖体大亚基 Ribosomal protein(LSU) | rpl2*,rpl14,rpl16*,rpl20,rpl22,rpl23,rpl32,rpl33,rpl36 | |
| 光合作用相关 Genes for photo- synthesis | ATP 合酶 ATP synthase | atpA,atpB,atpE,atpF*,atpH,atpI |
| 光和系统Ⅰ Photosystem Ⅰ | psaA,psaB,psaC,psaI,psaJ | |
| 光和系统Ⅱ Photosystem Ⅱ | psbA,psbB,psbC,psbD,psbE,psbF,psbI,psbJ,psbK,psbH,psbL,psbM,psbN,psbT,psbZ,ycf3** | |
| 二磷酸核酮糖羧化酶亚基 Subunit of rubisco | rbcL | |
| 细胞色素复合物 Cytochrome b/f complex | petA,petB*,petD*,petG,petL,petN | |
| NADH脱氢酶 NADH-dehydrogenase | ndhA*,ndhB*,ndhC,ndhD,ndhE,ndhF,ndhG,ndhH,ndhI,ndhJ,ndhK | |
| 其他与未知功能 Other and unknown function | accD,ccsA,cemA,clpP**,matK,ycf1,ycf2,ycf4,ycf15a |
| 基因 Gene | 正负链 Strand | 起始 Start | 终止 End | 外显子 Ⅰ/bp ExonⅠ | 内含子 Ⅰ/bp IntronⅠ | 外显子 Ⅱ/bp ExonⅡ | 内含子 Ⅱ/bp Intron Ⅱ | 外显子 Ⅲ/bp Exon Ⅲ | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | |||||||
| trnK-UUU | - | 1 607 | 1 607 | 1 607 | 4 237 | 4 237 | 4 237 | 37 | 2 559 | 35 | ||
| rps16 | - | 5 086 | 5 086 | 5 086 | 6 257 | 6 257 | 6 257 | 39 | 905 | 228 | ||
| trnG-GCC | + | 10 135 | 10 135 | 10 134 | 10 919 | 10 919 | 10 918 | 23 | 715 | 47 | ||
| atpF | - | 13 185 | 13 185 | 13 184 | 14 490 | 14 490 | 14 489 | 145 | 751 | 410 | ||
| rpoC1 | - | 22 810 | 22 810 | 22 811 | 25 681 | 25 681 | 25 683 | 432 | 814 | 1 626 | ||
| ycf3 | - | 47 097 | 47 097 | 47 034 | 49 116 | 49 116 | 49 053 | 124 | 720 | 230 | 793 | 153 |
| trnL-UAA | + | 52 205 | 52 205 | 52 142 | 52 815 | 52 815 | 52 752 | 37 | 524 | 50 | ||
| trnV-UAC | - | 56 413 | 56 413 | 56 350 | 57 102 | 57 102 | 57 039 | 38 | 615 | 37 | ||
| clpP | - | 75 722 | 75 722 | 75 665 | 77 773 | 77 773 | 77 715 | 71 | 847 | 294 | 614 | 226 |
| petB | + | 80 745 | 80 745 | 80 687 | 82 207 | 82 207 | 82 149 | 6 | 815 | 642 | ||
| petD | + | 82 418 | 82 418 | 82 360 | 83 526 | 83 526 | 83 468 | 9 | 626 | 474 | ||
| rpl16 | - | 87 056 | 87 056 | 86 998 | 88 382 | 88 382 | 88 324 | 9 | 919 | 399 | ||
| rpl2 | - | 90 103 | 90 103 | 90 045 | 91 625 | 91 625 | 91 567 | 391 | 698 | 434 | ||
| ndhB | - | 100 446 | 100 446 | 100 388 | 102 670 | 102 670 | 102 612 | 775 | 686 | 764 | ||
| trnI-GAU | + | 108 073 | 108 073 | 108 015 | 109 099 | 109 099 | 109 041 | 37 | 955 | 35 | ||
| trnA-UGC | + | 109 164 | 109 164 | 109 106 | 110 043 | 110 043 | 109 985 | 38 | 807 | 35 | ||
| ndhA | - | 125 970 | 127 762 | 125 912 | 128 253 | 130 045 | 128 195 | 553 | 1 192 | 539 | ||
| trnA-UGC | - | 140 799 | 142 591 | 140 742 | 141 678 | 143 470 | 141 621 | 38 | 807 | 35 | ||
| trnI-GAU | - | 141 743 | 143 535 | 141 686 | 142 769 | 144 561 | 142 712 | 37 | 955 | 35 | ||
| ndhB | + | 148 172 | 149 964 | 148 115 | 150 396 | 152 188 | 150 339 | 775 | 686 | 764 | ||
| rpl2 | + | 159 217 | 161 009 | 159 160 | 160 739 | 162 531 | 160 682 | 391 | 698 | 434 | ||
表3 薄壳山核桃‘金华’‘波尼’和实生单株叶绿体基因组中内含子和外显子长度
Table 3 Intron and exon length in the chloroplast genome of Carya illinoinensis‘Jinhua’‘Pawnee’and seedling individual
| 基因 Gene | 正负链 Strand | 起始 Start | 终止 End | 外显子 Ⅰ/bp ExonⅠ | 内含子 Ⅰ/bp IntronⅠ | 外显子 Ⅱ/bp ExonⅡ | 内含子 Ⅱ/bp Intron Ⅱ | 外显子 Ⅲ/bp Exon Ⅲ | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | |||||||
| trnK-UUU | - | 1 607 | 1 607 | 1 607 | 4 237 | 4 237 | 4 237 | 37 | 2 559 | 35 | ||
| rps16 | - | 5 086 | 5 086 | 5 086 | 6 257 | 6 257 | 6 257 | 39 | 905 | 228 | ||
| trnG-GCC | + | 10 135 | 10 135 | 10 134 | 10 919 | 10 919 | 10 918 | 23 | 715 | 47 | ||
| atpF | - | 13 185 | 13 185 | 13 184 | 14 490 | 14 490 | 14 489 | 145 | 751 | 410 | ||
| rpoC1 | - | 22 810 | 22 810 | 22 811 | 25 681 | 25 681 | 25 683 | 432 | 814 | 1 626 | ||
| ycf3 | - | 47 097 | 47 097 | 47 034 | 49 116 | 49 116 | 49 053 | 124 | 720 | 230 | 793 | 153 |
| trnL-UAA | + | 52 205 | 52 205 | 52 142 | 52 815 | 52 815 | 52 752 | 37 | 524 | 50 | ||
| trnV-UAC | - | 56 413 | 56 413 | 56 350 | 57 102 | 57 102 | 57 039 | 38 | 615 | 37 | ||
| clpP | - | 75 722 | 75 722 | 75 665 | 77 773 | 77 773 | 77 715 | 71 | 847 | 294 | 614 | 226 |
| petB | + | 80 745 | 80 745 | 80 687 | 82 207 | 82 207 | 82 149 | 6 | 815 | 642 | ||
| petD | + | 82 418 | 82 418 | 82 360 | 83 526 | 83 526 | 83 468 | 9 | 626 | 474 | ||
| rpl16 | - | 87 056 | 87 056 | 86 998 | 88 382 | 88 382 | 88 324 | 9 | 919 | 399 | ||
| rpl2 | - | 90 103 | 90 103 | 90 045 | 91 625 | 91 625 | 91 567 | 391 | 698 | 434 | ||
| ndhB | - | 100 446 | 100 446 | 100 388 | 102 670 | 102 670 | 102 612 | 775 | 686 | 764 | ||
| trnI-GAU | + | 108 073 | 108 073 | 108 015 | 109 099 | 109 099 | 109 041 | 37 | 955 | 35 | ||
| trnA-UGC | + | 109 164 | 109 164 | 109 106 | 110 043 | 110 043 | 109 985 | 38 | 807 | 35 | ||
| ndhA | - | 125 970 | 127 762 | 125 912 | 128 253 | 130 045 | 128 195 | 553 | 1 192 | 539 | ||
| trnA-UGC | - | 140 799 | 142 591 | 140 742 | 141 678 | 143 470 | 141 621 | 38 | 807 | 35 | ||
| trnI-GAU | - | 141 743 | 143 535 | 141 686 | 142 769 | 144 561 | 142 712 | 37 | 955 | 35 | ||
| ndhB | + | 148 172 | 149 964 | 148 115 | 150 396 | 152 188 | 150 339 | 775 | 686 | 764 | ||
| rpl2 | + | 159 217 | 161 009 | 159 160 | 160 739 | 162 531 | 160 682 | 391 | 698 | 434 | ||
| 密码子Codon | 氨基酸 Amino acid | 频率/% Frequency | 数量 Number | RSCU | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | ||
| GCA | A | 1.454 | 1.426 | 1.454 | 715 | 729 | 715 | 1.130 | 1.140 | 1.130 |
| GCC | A | 0.679 | 0.665 | 0.679 | 334 | 340 | 334 | 0.530 | 0.530 | 0.530 |
| GCG | A | 0.596 | 0.585 | 0.592 | 293 | 299 | 291 | 0.460 | 0.470 | 0.460 |
| GCT | A | 2.408 | 2.348 | 2.408 | 1 184 | 1 200 | 1 184 | 1.870 | 1.870 | 1.880 |
| TGC | C | 0.334 | 0.333 | 0.334 | 164 | 170 | 164 | 0.560 | 0.560 | 0.560 |
| TGT | C | 0.860 | 0.855 | 0.860 | 423 | 437 | 423 | 1.440 | 1.440 | 1.440 |
| GAC | D | 0.810 | 0.802 | 0.810 | 398 | 410 | 398 | 0.400 | 0.390 | 0.400 |
| GAT | D | 3.259 | 3.272 | 3.259 | 1 602 | 1 672 | 1 602 | 1.600 | 1.610 | 1.600 |
| GAA | E | 3.897 | 3.996 | 3.897 | 1 916 | 2 042 | 1 916 | 1.490 | 1.500 | 1.490 |
| GAG | E | 1.328 | 1.336 | 1.332 | 653 | 683 | 655 | 0.510 | 0.500 | 0.510 |
| TTC | F | 2.018 | 2.015 | 2.018 | 992 | 1 030 | 992 | 0.690 | 0.680 | 0.690 |
| TTT | F | 3.794 | 3.872 | 3.794 | 1 865 | 1 979 | 1 865 | 1.310 | 1.320 | 1.310 |
| GGA | G | 2.532 | 2.499 | 2.532 | 1 245 | 1 277 | 1 245 | 1.590 | 1.600 | 1.590 |
| GGC | G | 0.712 | 0.701 | 0.712 | 350 | 358 | 350 | 0.450 | 0.450 | 0.450 |
| GGG | G | 0.970 | 0.961 | 0.970 | 477 | 491 | 477 | 0.610 | 0.620 | 0.610 |
| GGT | G | 2.140 | 2.090 | 2.140 | 1 052 | 1 068 | 1 052 | 1.350 | 1.340 | 1.350 |
| CAC | H | 0.547 | 0.546 | 0.547 | 269 | 279 | 269 | 0.450 | 0.450 | 0.450 |
| CAT | H | 1.906 | 1.869 | 1.906 | 937 | 955 | 937 | 1.550 | 1.550 | 1.550 |
| ATA | I | 2.888 | 2.947 | 2.893 | 1 420 | 1 506 | 1 422 | 0.990 | 1.000 | 0.990 |
| ATC | I | 1.621 | 1.622 | 1.621 | 797 | 829 | 797 | 0.550 | 0.550 | 0.550 |
| ATT | I | 4.274 | 4.283 | 4.274 | 2 101 | 2 189 | 2 101 | 1.460 | 1.450 | 1.460 |
| AAA | K | 3.995 | 4.133 | 3.995 | 1 964 | 2 112 | 1 964 | 1.450 | 1.450 | 1.450 |
| AAG | K | 1.511 | 1.552 | 1.507 | 743 | 793 | 741 | 0.550 | 0.550 | 0.550 |
| CTA | L | 1.336 | 1.333 | 1.332 | 657 | 681 | 655 | 0.760 | 0.760 | 0.760 |
| CTC | L | 0.755 | 0.757 | 0.755 | 371 | 387 | 371 | 0.430 | 0.430 | 0.430 |
| CTG | L | 0.675 | 0.681 | 0.675 | 332 | 348 | 332 | 0.390 | 0.390 | 0.390 |
| CTT | L | 2.258 | 2.227 | 2.258 | 1 110 | 1 138 | 1 110 | 1.290 | 1.280 | 1.290 |
| TTA | L | 3.242 | 3.260 | 3.242 | 1 594 | 1 666 | 1 594 | 1.850 | 1.870 | 1.850 |
| TTG | L | 2.246 | 2.199 | 2.246 | 1 104 | 1 124 | 1 104 | 1.280 | 1.260 | 1.280 |
| ATG | M | 2.244 | 2.225 | 2.244 | 1 103 | 1 137 | 1 103 | 1.000 | 1.000 | 1.000 |
| AAC | N | 1.153 | 1.156 | 1.153 | 567 | 591 | 567 | 0.470 | 0.460 | 0.470 |
| AAT | N | 3.794 | 3.837 | 3.802 | 1 865 | 1 961 | 1 869 | 1.530 | 1.540 | 1.530 |
| 密码子Codon | 氨基酸 Amino acid | 频率/% Frequency | 数量 Number | RSCU | ||||||
| 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | ||
| CCA | P | 1.168 | 1.155 | 1.168 | 574 | 590 | 574 | 1.140 | 1.130 | 1.140 |
| CCC | P | 0.816 | 0.816 | 0.816 | 401 | 417 | 401 | 0.800 | 0.800 | 0.800 |
| CCG | P | 0.633 | 0.640 | 0.633 | 311 | 327 | 311 | 0.620 | 0.630 | 0.620 |
| CCT | P | 1.479 | 1.477 | 1.479 | 727 | 755 | 727 | 1.440 | 1.450 | 1.440 |
| CAA | Q | 2.805 | 2.820 | 2.805 | 1 379 | 1 441 | 1 379 | 1.550 | 1.560 | 1.550 |
| CAG | Q | 0.818 | 0.806 | 0.818 | 402 | 412 | 402 | 0.450 | 0.440 | 0.450 |
| AGA | R | 1.827 | 1.828 | 1.827 | 898 | 934 | 898 | 1.810 | 1.820 | 1.810 |
| AGG | R | 0.716 | 0.704 | 0.716 | 352 | 360 | 352 | 0.710 | 0.700 | 0.710 |
| CGA | R | 1.367 | 1.374 | 1.367 | 672 | 702 | 672 | 1.350 | 1.370 | 1.350 |
| CGC | R | 0.393 | 0.389 | 0.393 | 193 | 199 | 193 | 0.390 | 0.390 | 0.390 |
| CGG | R | 0.472 | 0.458 | 0.472 | 232 | 234 | 232 | 0.470 | 0.460 | 0.470 |
| CGT | R | 1.288 | 1.274 | 1.288 | 633 | 651 | 633 | 1.270 | 1.270 | 1.270 |
| AGC | S | 0.535 | 0.538 | 0.535 | 263 | 275 | 263 | 0.420 | 0.420 | 0.420 |
| AGT | S | 1.530 | 1.511 | 1.526 | 752 | 772 | 750 | 1.200 | 1.190 | 1.200 |
| TCA | S | 1.526 | 1.542 | 1.526 | 750 | 788 | 750 | 1.200 | 1.220 | 1.200 |
| TCC | S | 1.202 | 1.192 | 1.202 | 591 | 609 | 591 | 0.940 | 0.940 | 0.940 |
| TCG | S | 0.738 | 0.726 | 0.738 | 363 | 371 | 363 | 0.580 | 0.570 | 0.580 |
| TCT | S | 2.126 | 2.107 | 2.126 | 1 045 | 1 077 | 1 045 | 1.670 | 1.660 | 1.670 |
| ACA | T | 1.471 | 1.493 | 1.471 | 723 | 763 | 723 | 1.220 | 1.230 | 1.220 |
| ACC | T | 0.828 | 0.828 | 0.828 | 407 | 423 | 407 | 0.690 | 0.680 | 0.690 |
| ACG | T | 0.531 | 0.530 | 0.531 | 261 | 271 | 261 | 0.440 | 0.440 | 0.440 |
| ACT | T | 1.991 | 1.998 | 1.991 | 979 | 1 021 | 979 | 1.650 | 1.650 | 1.650 |
| GTA | V | 1.983 | 1.970 | 1.983 | 975 | 1 007 | 975 | 1.530 | 1.530 | 1.530 |
| GTC | V | 0.631 | 0.638 | 0.631 | 310 | 326 | 310 | 0.490 | 0.500 | 0.490 |
| GTG | V | 0.677 | 0.663 | 0.677 | 333 | 339 | 333 | 0.520 | 0.520 | 0.520 |
| GTT | V | 1.906 | 1.869 | 1.906 | 937 | 955 | 937 | 1.470 | 1.450 | 1.470 |
| TGG | W | 1.855 | 1.847 | 1.855 | 912 | 944 | 912 | 1.000 | 1.000 | 1.000 |
| TAC | Y | 0.767 | 0.753 | 0.767 | 377 | 385 | 377 | 0.410 | 0.400 | 0.410 |
| TAT | Y | 3.015 | 3.021 | 3.015 | 1 482 | 1 544 | 1 482 | 1.590 | 1.600 | 1.590 |
| TAA | * | 0.283 | 0.276 | 0.283 | 139 | 141 | 139 | 1.260 | 1.270 | 1.260 |
| TAG | * | 0.205 | 0.198 | 0.205 | 101 | 101 | 101 | 0.920 | 0.910 | 0.920 |
| TGA | * | 0.185 | 0.178 | 0.185 | 91 | 91 | 91 | 0.830 | 0.820 | 0.830 |
表4 薄壳山核桃‘金华’‘波尼’和实生单株叶绿体基因相对同义密码子使用度
Table 4 Relative synonymous codon usage in chloroplast genes of Carya illinoinensis‘Jinhua’‘Pawnee’and Seedling individual
| 密码子Codon | 氨基酸 Amino acid | 频率/% Frequency | 数量 Number | RSCU | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | ||
| GCA | A | 1.454 | 1.426 | 1.454 | 715 | 729 | 715 | 1.130 | 1.140 | 1.130 |
| GCC | A | 0.679 | 0.665 | 0.679 | 334 | 340 | 334 | 0.530 | 0.530 | 0.530 |
| GCG | A | 0.596 | 0.585 | 0.592 | 293 | 299 | 291 | 0.460 | 0.470 | 0.460 |
| GCT | A | 2.408 | 2.348 | 2.408 | 1 184 | 1 200 | 1 184 | 1.870 | 1.870 | 1.880 |
| TGC | C | 0.334 | 0.333 | 0.334 | 164 | 170 | 164 | 0.560 | 0.560 | 0.560 |
| TGT | C | 0.860 | 0.855 | 0.860 | 423 | 437 | 423 | 1.440 | 1.440 | 1.440 |
| GAC | D | 0.810 | 0.802 | 0.810 | 398 | 410 | 398 | 0.400 | 0.390 | 0.400 |
| GAT | D | 3.259 | 3.272 | 3.259 | 1 602 | 1 672 | 1 602 | 1.600 | 1.610 | 1.600 |
| GAA | E | 3.897 | 3.996 | 3.897 | 1 916 | 2 042 | 1 916 | 1.490 | 1.500 | 1.490 |
| GAG | E | 1.328 | 1.336 | 1.332 | 653 | 683 | 655 | 0.510 | 0.500 | 0.510 |
| TTC | F | 2.018 | 2.015 | 2.018 | 992 | 1 030 | 992 | 0.690 | 0.680 | 0.690 |
| TTT | F | 3.794 | 3.872 | 3.794 | 1 865 | 1 979 | 1 865 | 1.310 | 1.320 | 1.310 |
| GGA | G | 2.532 | 2.499 | 2.532 | 1 245 | 1 277 | 1 245 | 1.590 | 1.600 | 1.590 |
| GGC | G | 0.712 | 0.701 | 0.712 | 350 | 358 | 350 | 0.450 | 0.450 | 0.450 |
| GGG | G | 0.970 | 0.961 | 0.970 | 477 | 491 | 477 | 0.610 | 0.620 | 0.610 |
| GGT | G | 2.140 | 2.090 | 2.140 | 1 052 | 1 068 | 1 052 | 1.350 | 1.340 | 1.350 |
| CAC | H | 0.547 | 0.546 | 0.547 | 269 | 279 | 269 | 0.450 | 0.450 | 0.450 |
| CAT | H | 1.906 | 1.869 | 1.906 | 937 | 955 | 937 | 1.550 | 1.550 | 1.550 |
| ATA | I | 2.888 | 2.947 | 2.893 | 1 420 | 1 506 | 1 422 | 0.990 | 1.000 | 0.990 |
| ATC | I | 1.621 | 1.622 | 1.621 | 797 | 829 | 797 | 0.550 | 0.550 | 0.550 |
| ATT | I | 4.274 | 4.283 | 4.274 | 2 101 | 2 189 | 2 101 | 1.460 | 1.450 | 1.460 |
| AAA | K | 3.995 | 4.133 | 3.995 | 1 964 | 2 112 | 1 964 | 1.450 | 1.450 | 1.450 |
| AAG | K | 1.511 | 1.552 | 1.507 | 743 | 793 | 741 | 0.550 | 0.550 | 0.550 |
| CTA | L | 1.336 | 1.333 | 1.332 | 657 | 681 | 655 | 0.760 | 0.760 | 0.760 |
| CTC | L | 0.755 | 0.757 | 0.755 | 371 | 387 | 371 | 0.430 | 0.430 | 0.430 |
| CTG | L | 0.675 | 0.681 | 0.675 | 332 | 348 | 332 | 0.390 | 0.390 | 0.390 |
| CTT | L | 2.258 | 2.227 | 2.258 | 1 110 | 1 138 | 1 110 | 1.290 | 1.280 | 1.290 |
| TTA | L | 3.242 | 3.260 | 3.242 | 1 594 | 1 666 | 1 594 | 1.850 | 1.870 | 1.850 |
| TTG | L | 2.246 | 2.199 | 2.246 | 1 104 | 1 124 | 1 104 | 1.280 | 1.260 | 1.280 |
| ATG | M | 2.244 | 2.225 | 2.244 | 1 103 | 1 137 | 1 103 | 1.000 | 1.000 | 1.000 |
| AAC | N | 1.153 | 1.156 | 1.153 | 567 | 591 | 567 | 0.470 | 0.460 | 0.470 |
| AAT | N | 3.794 | 3.837 | 3.802 | 1 865 | 1 961 | 1 869 | 1.530 | 1.540 | 1.530 |
| 密码子Codon | 氨基酸 Amino acid | 频率/% Frequency | 数量 Number | RSCU | ||||||
| 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | 金华 Jinhua | 波尼 Pawnee | 实生单株 Seedling individual | ||
| CCA | P | 1.168 | 1.155 | 1.168 | 574 | 590 | 574 | 1.140 | 1.130 | 1.140 |
| CCC | P | 0.816 | 0.816 | 0.816 | 401 | 417 | 401 | 0.800 | 0.800 | 0.800 |
| CCG | P | 0.633 | 0.640 | 0.633 | 311 | 327 | 311 | 0.620 | 0.630 | 0.620 |
| CCT | P | 1.479 | 1.477 | 1.479 | 727 | 755 | 727 | 1.440 | 1.450 | 1.440 |
| CAA | Q | 2.805 | 2.820 | 2.805 | 1 379 | 1 441 | 1 379 | 1.550 | 1.560 | 1.550 |
| CAG | Q | 0.818 | 0.806 | 0.818 | 402 | 412 | 402 | 0.450 | 0.440 | 0.450 |
| AGA | R | 1.827 | 1.828 | 1.827 | 898 | 934 | 898 | 1.810 | 1.820 | 1.810 |
| AGG | R | 0.716 | 0.704 | 0.716 | 352 | 360 | 352 | 0.710 | 0.700 | 0.710 |
| CGA | R | 1.367 | 1.374 | 1.367 | 672 | 702 | 672 | 1.350 | 1.370 | 1.350 |
| CGC | R | 0.393 | 0.389 | 0.393 | 193 | 199 | 193 | 0.390 | 0.390 | 0.390 |
| CGG | R | 0.472 | 0.458 | 0.472 | 232 | 234 | 232 | 0.470 | 0.460 | 0.470 |
| CGT | R | 1.288 | 1.274 | 1.288 | 633 | 651 | 633 | 1.270 | 1.270 | 1.270 |
| AGC | S | 0.535 | 0.538 | 0.535 | 263 | 275 | 263 | 0.420 | 0.420 | 0.420 |
| AGT | S | 1.530 | 1.511 | 1.526 | 752 | 772 | 750 | 1.200 | 1.190 | 1.200 |
| TCA | S | 1.526 | 1.542 | 1.526 | 750 | 788 | 750 | 1.200 | 1.220 | 1.200 |
| TCC | S | 1.202 | 1.192 | 1.202 | 591 | 609 | 591 | 0.940 | 0.940 | 0.940 |
| TCG | S | 0.738 | 0.726 | 0.738 | 363 | 371 | 363 | 0.580 | 0.570 | 0.580 |
| TCT | S | 2.126 | 2.107 | 2.126 | 1 045 | 1 077 | 1 045 | 1.670 | 1.660 | 1.670 |
| ACA | T | 1.471 | 1.493 | 1.471 | 723 | 763 | 723 | 1.220 | 1.230 | 1.220 |
| ACC | T | 0.828 | 0.828 | 0.828 | 407 | 423 | 407 | 0.690 | 0.680 | 0.690 |
| ACG | T | 0.531 | 0.530 | 0.531 | 261 | 271 | 261 | 0.440 | 0.440 | 0.440 |
| ACT | T | 1.991 | 1.998 | 1.991 | 979 | 1 021 | 979 | 1.650 | 1.650 | 1.650 |
| GTA | V | 1.983 | 1.970 | 1.983 | 975 | 1 007 | 975 | 1.530 | 1.530 | 1.530 |
| GTC | V | 0.631 | 0.638 | 0.631 | 310 | 326 | 310 | 0.490 | 0.500 | 0.490 |
| GTG | V | 0.677 | 0.663 | 0.677 | 333 | 339 | 333 | 0.520 | 0.520 | 0.520 |
| GTT | V | 1.906 | 1.869 | 1.906 | 937 | 955 | 937 | 1.470 | 1.450 | 1.470 |
| TGG | W | 1.855 | 1.847 | 1.855 | 912 | 944 | 912 | 1.000 | 1.000 | 1.000 |
| TAC | Y | 0.767 | 0.753 | 0.767 | 377 | 385 | 377 | 0.410 | 0.400 | 0.410 |
| TAT | Y | 3.015 | 3.021 | 3.015 | 1 482 | 1 544 | 1 482 | 1.590 | 1.600 | 1.590 |
| TAA | * | 0.283 | 0.276 | 0.283 | 139 | 141 | 139 | 1.260 | 1.270 | 1.260 |
| TAG | * | 0.205 | 0.198 | 0.205 | 101 | 101 | 101 | 0.920 | 0.910 | 0.920 |
| TGA | * | 0.185 | 0.178 | 0.185 | 91 | 91 | 91 | 0.830 | 0.820 | 0.830 |
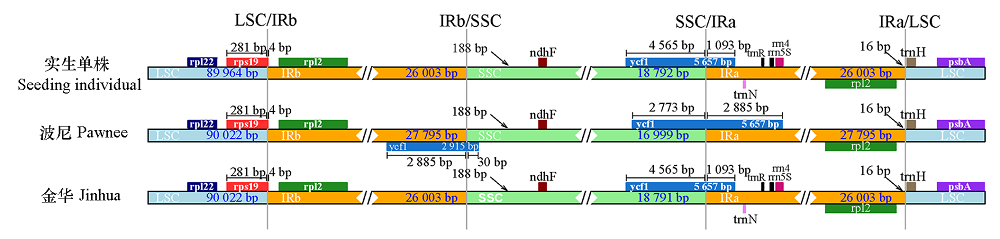
图3 薄壳山核桃‘金华’‘波尼’和实生单株叶绿体基因组LSC、SSC和IR区边界的比较
Fig. 3 Comparison of LSC,SSC and IR boundary of chloroplast genomes of Carya illinoinensis‘Jinhua’‘Pawnee’and seedling individual
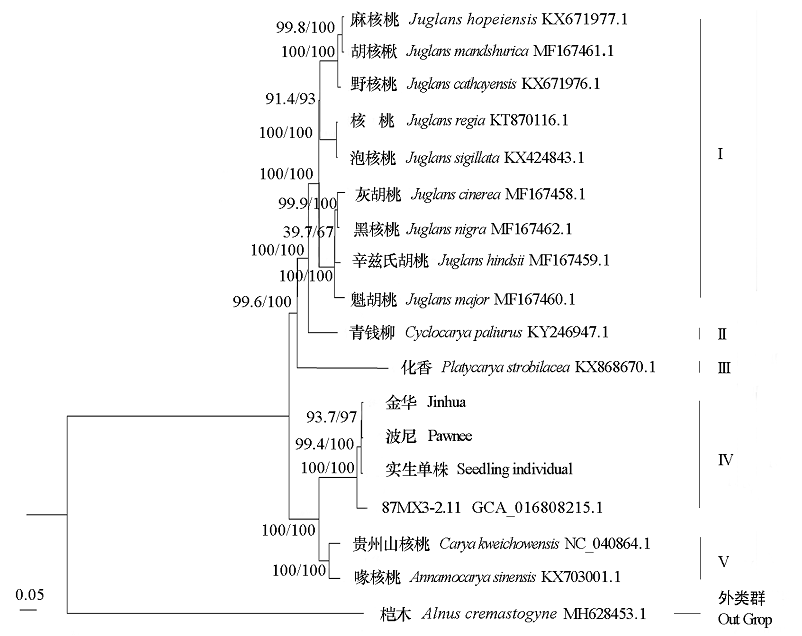
图4 基于叶绿体基因组串联序列构建18个物种的最大似然(ML)系统进化树
Fig. 4 Maximum likelihood(ML)phylogenetic tree construction including 18 species based on concatenated sequences from all chloroplast genomes
| [1] | Alexander L W, Woeste K E. 2014. Pyrosequencing of the northern red oak(Quercus rubra L.)chloroplast genome reveals high quality polymorphisms for population management. Tree Genetics & Genomes, 10 (4):803-812. |
| [2] |
Beier S, Thiel T, Munch T, Scholz U, Mascher M. 2017. MISA-web:a web server for microsatellite prediction. Bioinformatics, 33:2583-2585.
doi: 10.1093/bioinformatics/btx198 URL |
| [3] | Chan P P, Lowe T M. 2019. tRNAscan-SE:searching for tRNA genes in genomic sequences. Methods in Molecular Biology, 1962:1-14. |
| [4] | Chang Jun, Zhang Xiao-dan, Yao Xiao-hua, Yang Shui-ping, Wang Kai-liang, Ren Hua-dong. 2021. Amino acid composition and nutritional value evaluation of different varieties of pecan(Carya illinoensis K. Koch). Journal of Southwest University(Natural Science Edition), 43 (4):44-52. (in Chinese) |
| 常君, 张潇丹, 姚小华, 杨水平, 王开良, 任华东. 2021. 不同品种薄壳山核桃氨基酸组成及营养价值评价. 西南大学学报(自然科学版), 43 (4):44-52. | |
| [5] | Dong W, Xu C, Cheng T, Zhou S. 2013. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE, 8 (10):e77965. |
| [6] | Frazer K A, Lior P, Alexander P, Rubin E M, Inna D. 2004. VISTA:Computational tools for comparative genomics. Nucleic Acids Research, 32:W273-W279. |
| [7] | Grauke L J, Mendoza-herrera M A, Binzel M L. 2010. Plastid microsatellite markers in Carya. Acta Horticulturae, 859:237-246. |
| [8] |
Grauke L J, Wood B W, Harris M K. 2016. Crop vulnerability:Carya. HortScience, 51 (6):653-663.
doi: 10.21273/HORTSCI.51.6.653 URL |
| [9] |
Greiner S, Lehwark P, Boc R. 2019. OrganellarGenomeDRAW(OGDRAW)version 1.3.1:expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research, 47:W59-W64.
doi: 10.1093/nar/gkz238 URL |
| [10] | Hou Dong-pei, Xi Xue-liang, Shi Zhuo-gong. 2007. The survey research of Carya illinoensis in China. Journal of Shandong Forestry Science and Technology,(4):53-55. (in Chinese) |
| 侯冬培, 习学良, 石卓功. 2007. 我国薄壳山核桃研究概况. 山东林业科技,(4):53-55. | |
| [11] | Hu Yi-heng. 2018. Phylogenetic and population genetics of Juglandaceae based on genomics and transcriptomics[M. D. Dissertation]. Xi’an: Northwestern University. (in Chinese) |
| 胡昳恒. 2018. 基于基因组学与转录组学的胡桃科植物系统进化及群体遗传学研究[硕士论文]. 西安: 西北大学. | |
| [12] |
Ishizuka W, Tabata A, Ono K, Fukuda Y, Hara T. 2017. Draft chloroplast genome of Larix gmelinii var. japonica:insight into intraspecific divergence. Journal of Forest Research, 22 (6):393-398.
doi: 10.1080/13416979.2017.1386019 URL |
| [13] | Jansen R K, Cai Z, Raubeson L A, Daniell H, Depamphilis C W, Leebens-Mack J, Müller K F, Guisinger-Bellian M, Haberle R C, Hansen A K, Chumley T W, Lee S B, Peery R, McNeal J R, Kuehl J V, Boore J L. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences, 104 (49):19369-19374. |
| [14] |
Jansen R K, Kaittanis C, Saski C, Lee S B, Tomkins J, Alverson A J, Daniell H. 2006. Phylogenetic analyses of Vitis(Vitaceae)based on complete chloroplast genome sequences:effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evolutionary Biology, 6 (1):1-14.
doi: 10.1186/1471-2148-6-1 URL |
| [15] |
Kang H I, Lee H O, Lee I H, Kim I S, Shim D. 2019. Complete chloroplast genome of Pinus densiflora Siebold & Zucc. and comparative analysis with five pine trees. Forests, 10 (7):600.
doi: 10.3390/f10070600 URL |
| [16] | Li Qian, Guo Qiqiang, Gao Chao, Li Hui’e. 2020. Characterization of complete chloroplast genome of Camellia weiningensis in Weining,Guizhou Province. Acta Horticulturae Sinica, 47 (4):779-781. (in Chinese) |
| 李倩, 郭其强, 高超, 李慧娥. 2020. 贵州威宁红花油茶的叶绿体基因组特征分析. 园艺学报, 47 (4):779-787. | |
| [17] | Li Yongtan, Zhang Jun, Huang Yali, Fan Jianmin, Zhang Yiwen, Zuo Lihui. 2020. Analysis of chloroplast genome of Pyrus betulaefolia. Acta Horticulturae Sinica, 47 (6):1021-1032. (in Chinese) |
| 李泳潭, 张军, 黄亚丽, 范建敏, 张益文, 左力辉. 2020. 杜梨叶绿体基因组分析. 园艺学报, 47 (6):1021-1032. | |
| [18] |
Lin C P, Wu C S, Huang Y Y, Chaw S M. 2012. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biology and Evolution, 4 (3):374-381.
doi: 10.1093/gbe/evs021 URL |
| [19] |
Lin C S, Chen J J, Chiu C C, Hsiao H C, Yang C J, Jin X H, Leebens-Mack J, de Pamphilis C W, Huang Y T, Yang L H, Chang W J, Kui L W, Gane K S H, Wang J M, WenShih M C. 2017. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genome is some orchids. The Plant Journal, 90 (5):994-1006.
doi: 10.1111/tpj.13525 URL |
| [20] |
Liu Q, Xue Q. 2005. Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. Journal of Genetics, 84 (1):55-62.
doi: 10.1007/BF02715890 URL |
| [21] |
Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research, 41:W575-W581.
doi: 10.1093/nar/gkt289 URL |
| [22] | Luo R B, Liu B H, Xie Y L, Li Z Y, Huang W H, Yuan J Y, He G Z, Chen Y X, Pan Q, Liu Y J, Tang J B, Wu G X, Zhuang H, Shi Y J, Liu Y, Yu C, Wang B, Lu Y, Han C L, Cheung D W, Yiu S M, Peng S L, Zhu X Q, Liu G M, Liao X K, Li Y R, Yang H M, Wang J, Lam W T, Wang J. 2012. SOAPdenovo2:an empirically improved memory-efficient short-read de novo assembler. GigaScience, 1 (1):2047-217X-1-18. |
| [23] |
Manos P S, Soltis P S, Soltis D E, Manchester S R, Oh S H, Bel C D, Dilcher D L, Stone D E. 2007. Phylogeny of extant and fossil Juglandaceae inferred from the integration of molecular and morphological data sets. Systematic Biology, 56 (3):412-430.
pmid: 17558964 |
| [24] |
Manos P S, Stone D E. 2001. Evolution,phylogeny,and systematics of the Juglandaceae. Annals of the Missouri Botanical Garden, 88 (2):231-269.
doi: 10.2307/2666226 URL |
| [25] |
Mo Z, Lou W, Chen Y, Jia X, Zhai M, Guo Z, Xuan J. 2020. The chloroplast genome of Carya illinoinensis:genome structure,adaptive evolution,and phylogenetic analysis. Forests, 11 (2):207.
doi: 10.3390/f11020207 URL |
| [26] | Moore M J, Soltis P S, Bell C D, Burleigh J G, Soltis D E. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proceedings of the National Academy of Sciences, 107 (10):4623-4628. |
| [27] | Olsson S, Grivet D, Vian J C. 2018. Species-diagnostic markers in the genus Pinus:evaluation of the chloroplast regions matK and ycf1. Forest Systems, 27 (3):2. |
| [28] |
Palmer J D, Thompson W F. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell, 29 (2):537-550.
pmid: 6288261 |
| [29] | Qi Jian-xun, Hao Yan-bin, Zhu Yan, Wu Chun-lin, Wang Wei-xia, Leng Ping. 2011. Studies on germplasm of Juglans by EST-SSR markers. Acta Horticulturae Sinica, 38 (3):441-448. (in Chinese) |
| 齐建勋, 郝艳宾, 朱艳, 吴春林, 王维霞, 冷平. 2011. 核桃属种质资源的EST-SSR标记研究. 园艺学报, 38 (3):441-448. | |
| [30] |
Raman G, Park S J. 2016. The complete chloroplast genome sequence of ampelopsis:gene organization,comparative analysis,and phylogenetic relationships to other angiosperms. Frontiers in Plant Science, 7:341.
doi: 10.3389/fpls.2016.00341 URL |
| [31] |
Ranade S S, Garcia-Gil M R, Rossello J A. 2016. Non-functional plastid ndh gene fragments are present in the nuclear genome of Norway spruce (Picea abies L. Karsch):insights from in silico analysis of nuclear and organellar genomes. Molecular Genetics and Genomics, 291 (2):935-941.
doi: 10.1007/s00438-015-1159-7 URL |
| [32] |
Redwan R M, Saidin A, Kumar S V. 2015. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biology, 15 (196):1-20.
doi: 10.1186/s12870-014-0410-4 URL |
| [33] |
Sharp P M, Li W H. 1987. The codon adaptation index-a measure of directional synonymous codon usage bias,and its potential applications. Nucleic Acids Research, 15 (3):1281-1295.
pmid: 3547335 |
| [34] |
Stamatakis A. 2014. RAxML version 8:a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30 (9):1312-1313.
doi: 10.1093/bioinformatics/btu033 URL |
| [35] |
Sugiura M. 1992. The chloroplast genome. Plant Molecular Biology, 19 (1):149-168.
pmid: 1600166 |
| [36] | Thompson T E, Grauke L J. 2000. ‘Pawnee’pecan. Journal of American Pomological Society, 54 (3):110-113. |
| [37] | Wang Man, Ning De-lu, Li Xian-zhong, Zhang Yu, Li Yong-jie. 2010. The survey research and development trends of Carya illinoensis. Forest by-Product and Speciality in China,(2):84-86. (in Chinese) |
| 王曼, 宁德鲁, 李贤忠, 张雨, 李勇杰. 2010. 薄壳山核桃研究概况. 中国林副特产,(2):84-86. | |
| [38] |
Wu F, Li M, Liao B, Shi X, Xu Y. 2019. DNA barcoding analysis and phylogenetic relation of mangroves in Guangdong Province,China. Forests, 10 (1):56.
doi: 10.3390/f10010056 URL |
| [39] | Wu Yan-min, Liu Ying, Dong Feng-xiang, Xi Sheng-ke. 2000a. Study on different ecological types of Chinese walnut(J. regia)using RAPD marker. Journal of Beijing Forestry University, 22 (5):23-27. (in Chinese) |
| 吴燕民, 刘英, 董凤祥, 奚声珂. 2000a. 应用RAPD对我国栽培核桃不同地理生态型的研究. 北京林业大学学报, 22 (5):23-27. | |
| [40] | Wu Yan-min, Pei Dong, Xi Sheng-ke, Li Jia-rui. 2000b. A study on the genetic relationship among species in Juglans L. using RAPD marker. Acta Horticulturae Sinica, 27 (1):17-22. (in Chinese) |
| 吴燕民, 裴东, 奚声珂, 李嘉瑞. 2000b. 运用RAPD对核桃属种间亲缘关系的研究. 园艺学报, 27 (1):17-22. | |
| [41] |
Xiang X G, Wang W, Li R Q, Lin L, Liu Y, Zhou Z K, Li Z Y, Chen Z D. 2014. Large-scale phylogenetic analyses reveal fagalean diversification promoted by the interplay of diaspores and environments in the Paleogene. Perspectives in Plant Ecology,Evolution and Systematics, 16 (3):101-110.
doi: 10.1016/j.ppees.2014.03.001 URL |
| [42] |
Xie H, Jiao J, Fan X, Zhang Y, Jiang J, Liu C. 2016. The complete chloroplast genome sequence of Chinese wild grape Vitis amurensis (Vitaceae:Vitis L.). Conservation Genetics Resources, 9 (1):1-4.
doi: 10.1007/s12686-016-0602-3 URL |
| [43] |
Yan C, Du J, Gao L, Li Y, Hou X. 2019. The complete chloroplast genome sequence of watercress(Nasturtium officinale R. Br.):genome organization,adaptive evolution and phylogenetic relationships in Cardamineae. Gene, 699:24-36.
doi: 10.1016/j.gene.2019.02.075 URL |
| [44] | Yang Jian-hua, Xi Xue-liang, Dong Run-quan, Fan Zhi-yuan, Li Shu-fang, Zou Wei-lie, Xiong Xin-wu, Chen Qin. 2018. Breeding of thin-shished hickory variety‘Jinhua’. South China Fruits, 47 (1):149-150. (in Chinese) |
| 杨建华, 习学良, 董润泉, 范志远, 李淑芳, 邹伟烈, 熊新武, 陈勤. 2018. 薄壳山核桃品种‘金华’的选育. 中国南方果树, 47 (1):149-150. | |
| [45] | Yang Y, Dong Y Y, Li Q, Lu J J, Li X W, Wang Y T. 2014. Complete chloroplast genome sequence of poisonous and medicinal plant Datura stramonium:organizations and implications for genetic engineering. PLoS ONE, 9 (11):e110656. |
| [46] | Yang Yameng, Jiao Jian, Fan Xiucai, Zhang Ying, Jiang Jianfu, Li Min, Liu Chonghuai. 2019. Complete chloroplast genome sequence and characteristics analysis of Vitis ficifolia. Acta Horticulturae Sinica, 46 (4):635-648. (in Chinese) |
|
杨亚蒙, 焦健, 樊秀彩, 张颖, 姜建福, 李民, 刘崇怀. 2019. 桑叶葡萄叶绿体基因组及其特征分析. 园艺学报, 46 (4):635-648.
doi: 10.16420/j.issn.0513-353x.2018-0596 |
|
| [47] |
Zhang R, Peng F, Li Y. 2015. Pecan production in China. Scientia Horticulturae, 197:719-727.
doi: 10.1016/j.scienta.2015.10.035 URL |
| [48] | Zheng Yi, Zhang Hui, Wang Qinmei, Gao Yue, Zhang Zhihong, Sun Yuxin. 2020. Complete chloroplast genome sequence of Clivia miniata and its characteristics. Acta Horticulturae Sinica, 47 (12):2439-2450. (in Chinese) |
| 郑祎, 张卉, 王钦美, 高悦, 张志宏, 孙玉新. 2020. 大花君子兰叶绿体基因组及其特征. 园艺学报, 47 (12):2439-2450. | |
| [49] | Zuo L H, Shang A Q, Zhang S, Yu X Y, Ren Y C, Yang M S, Wang J M. 2017. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing:Genome comparative and taxonomic position analysis. PLoS ONE, 12 (2):e0171264. |
| [1] | 季琳琳, 陈素传, 吴志辉, 常 君, 韩文妍, 陶汝鹏. 早花山核桃新品种‘宁国山核桃2号’[J]. 园艺学报, 2022, 49(S2): 53-54. |
| [2] | 丁志杰, 包金波, 柔鲜古丽, 朱甜甜, 李雪丽, 苗浩宇, 田新民. 新疆野苹果与‘元帅’‘金冠’的叶绿体基因组比对研究[J]. 园艺学报, 2022, 49(9): 1977-1990. |
| [3] | 汤晨茜, 仇志欣, 檀超, 钱羽铭, 陈昕. 陕甘花楸叶绿体基因组及其与爪瓣花楸的系统关系[J]. 园艺学报, 2022, 49(3): 641-654. |
| [4] | 宋芸, 贾孟君, 曹亚萍, 李政, 贺嘉欣, 王勇飞, 张鑫瑞, 乔永刚. 连翘叶绿体基因组特征分析[J]. 园艺学报, 2022, 49(1): 187-199. |
| [5] | 陈素传, 季琳琳, 吴志辉, 常 君, 陶汝鹏, 汪小进 . 大果山核桃新品种‘宁国山核桃3号’[J]. 园艺学报, 2021, 48(S2): 2803-2804. |
| [6] | 常 君, 姚小华, 王开良, 滕建华, 邵慰忠, 张建忠, 李雪涛, 任华东, . 薄壳山核桃新品种‘亚优YLC21号’[J]. 园艺学报, 2021, 48(S2): 2805-2806. |
| [7] | 陈素传, 吴志辉, 季琳琳, 常 君, 陶汝鹏, 汪小进. 山核桃新品种‘宁国山核桃1号’[J]. 园艺学报, 2020, 47(S2): 2925-2926. |
| [8] | 王开良, 姚小华, 邵慰忠, 任华东, 常 君, 方新高, 洪友君. 薄壳山核桃新品种‘亚优YLC28号’[J]. 园艺学报, 2020, 47(S2): 2927-2928. |
| [9] | 李泳潭,张 军*,黄亚丽,范建敏,张益文,左力辉. 杜梨叶绿体基因组分析[J]. 园艺学报, 2020, 47(6): 1021-1032. |
| [10] | 李 倩1,郭其强2,高 超2,李慧娥1,*. 贵州威宁红花油茶的叶绿体基因组特征分析[J]. 园艺学报, 2020, 47(4): 779-787. |
| [11] | 郑 祎, 张 卉, 王钦美, 高 悦, 张志宏, 孙玉新. 大花君子兰叶绿体基因组及其特征[J]. 园艺学报, 2020, 47(12): 2439-2450. |
| [12] | 杨亚蒙1,焦 健2,樊秀彩1,张 颖1,姜建福1,李 民1,刘崇怀1,*. 桑叶葡萄叶绿体基因组及其特征分析[J]. 园艺学报, 2019, 46(4): 635-648. |
| [13] | 朱先富1,何国庆2,黄久奎3,徐步青4,夏国华5,*. 大别山山核桃新品种‘皖金1号’[J]. 园艺学报, 2018, 45(S2): 2731-2732. |
| [14] | 夏国华1,王正加1,丁立忠2,张秋月3,黄坚钦1,*. 山核桃新品种‘浙林山3号’[J]. 园艺学报, 2018, 45(3): 603-604. |
| [15] | 黄春颖,黄有军,吴建峰,黄 仁,栾雨濛,张深梅,王正加,张启香,黄坚钦*. SAD和FAD家族基因调控山核桃不饱和脂肪酸组分配比[J]. 园艺学报, 2018, 45(2): 250-260. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司