
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (5): 1013-1022.doi: 10.16420/j.issn.0513-353x.2020-0497
• Research Notes • Previous Articles Next Articles
JIANG Shuang*( ), ZHANG Xueying, AN Haishan, XU Fangjie, ZHANG Jiaying
), ZHANG Xueying, AN Haishan, XU Fangjie, ZHANG Jiaying
Received:2020-12-15
Revised:2021-02-08
Online:2021-05-25
Published:2021-06-07
CLC Number:
JIANG Shuang, ZHANG Xueying, AN Haishan, XU Fangjie, ZHANG Jiaying. Development and Analysis of Polymorphism of SSR Markers in the Whole Genome of Loquat[J]. Acta Horticulturae Sinica, 2021, 48(5): 1013-1022.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0497
| 标记 Marker | 位置 Position | 上游引物(5′-3′) F-primer | 下游引物(5′-3′) R-primer |
|---|---|---|---|
| EjSSR1 | LG06_22780531_22780756 | GTGGCGGATACAAGATTTCA | CACCATTTGCCATTCTACCG |
| EjSSR2 | LG07_28161826_28162057 | GGCAAGTTTGTCACGCTATT | AAGAAAAGAAAGCGGTAATCGT |
| EjSSR3 | LG02_12437057_12437286 | TATGCAGCCATACTAATGTA | TTAGCGTAGACTCCAGGTGA |
| EjSSR4 | LG15_2449146_2449395 | ACAAGATTATGCAGCTACCTTAC | CCGCTAACGATGGAATGAAC |
| EjSSR5 | LG17_10249126_10249370 | GAGGGTCTGTTGGCTTGGAT | GAAATGTGGGCATCTTCTTC |
| EjSSR6 | LG02_13068397_13068630 | AATATGAGACGTACAAGCTG | TCTTTACTCGCACTGAATGT |
| EjSSR7 | LG09_33605598_33605838 | AAATGATCCCTCAACTATAACC | CATTGCTCTATACTCGCATC |
| EjSSR8 | LG05_8474062_8474283 | ATAAATAGCAGCAAGGGCTCCAT | GAAACCCCTACCTCAAATCCAA |
| EjSSR9 | LG16_306060_306291 | TTCGTACTCATTGAGTTTGGTAT | TGTTGATTAGAATACTGCCTCCC |
| EjSSR10 | LG05_7308571_7308855 | CCCATAATCCATCTAAGTACCAAA | GACCTTGAAGGACCTCCATACAC |
| EjSSR11 | LG04_14003826_14004069 | CTCGGAGGATGATAAATCAATTC | CCTATTCGACAAGGACGACAAT |
| EjSSR12 | LG11_26083798_26084019 | GCCTAAACTTGCCTAGATTCTGAC | ACGGAGCCTAAGCGGACTAA |
| EjSSR13 | LG07_37064315_37064546 | TTGGACGGGTTGAGGAACTTAG | CCCACGACTTTGTCTCGCTTCT |
| EjSSR14 | LG17_32809624_32809843 | TGCAGTGCCGAACTAAGCTACATA | ACCTGCTGTACCGCAACTTCTG |
| EjSSR15 | LG06_27714710_27714930 | ATTGGGAAGCCGTAGTTGGTGA | TCGCAGCTCTTTCTTCTCACTT |
| EjSSR16 | LG01_38250590_38250807 | AGTTGAGGCTTAGAAGGCAAGG | GAAAGAACTTTGAAAATTCCATCA |
| EjSSR17 | LG12_5320901_5321127 | GAGTGGGACTTCGCATTTGATA | GGAATAACTAACGCCCGCAAC |
| EjSSR18 | LG03_45350948_45351163 | CTCGTTATTGCGTTCAAGCATATT | TTTCTGTTTGGAGCTGGGATTT |
| EjSSR19 | LG16_264901_265127 | CCTTGCCCTGTAGTATTATGTGAT | ATCTGCGGATAAGGAGCACAAG |
| EjSSR20 | LG14_23903168_23903387 | TTTGCGTGATGATCGTTGTCCT | ACACTAAACGGGAGGATGTGGG |
Table 1 The list of tested SSR markers and their primers
| 标记 Marker | 位置 Position | 上游引物(5′-3′) F-primer | 下游引物(5′-3′) R-primer |
|---|---|---|---|
| EjSSR1 | LG06_22780531_22780756 | GTGGCGGATACAAGATTTCA | CACCATTTGCCATTCTACCG |
| EjSSR2 | LG07_28161826_28162057 | GGCAAGTTTGTCACGCTATT | AAGAAAAGAAAGCGGTAATCGT |
| EjSSR3 | LG02_12437057_12437286 | TATGCAGCCATACTAATGTA | TTAGCGTAGACTCCAGGTGA |
| EjSSR4 | LG15_2449146_2449395 | ACAAGATTATGCAGCTACCTTAC | CCGCTAACGATGGAATGAAC |
| EjSSR5 | LG17_10249126_10249370 | GAGGGTCTGTTGGCTTGGAT | GAAATGTGGGCATCTTCTTC |
| EjSSR6 | LG02_13068397_13068630 | AATATGAGACGTACAAGCTG | TCTTTACTCGCACTGAATGT |
| EjSSR7 | LG09_33605598_33605838 | AAATGATCCCTCAACTATAACC | CATTGCTCTATACTCGCATC |
| EjSSR8 | LG05_8474062_8474283 | ATAAATAGCAGCAAGGGCTCCAT | GAAACCCCTACCTCAAATCCAA |
| EjSSR9 | LG16_306060_306291 | TTCGTACTCATTGAGTTTGGTAT | TGTTGATTAGAATACTGCCTCCC |
| EjSSR10 | LG05_7308571_7308855 | CCCATAATCCATCTAAGTACCAAA | GACCTTGAAGGACCTCCATACAC |
| EjSSR11 | LG04_14003826_14004069 | CTCGGAGGATGATAAATCAATTC | CCTATTCGACAAGGACGACAAT |
| EjSSR12 | LG11_26083798_26084019 | GCCTAAACTTGCCTAGATTCTGAC | ACGGAGCCTAAGCGGACTAA |
| EjSSR13 | LG07_37064315_37064546 | TTGGACGGGTTGAGGAACTTAG | CCCACGACTTTGTCTCGCTTCT |
| EjSSR14 | LG17_32809624_32809843 | TGCAGTGCCGAACTAAGCTACATA | ACCTGCTGTACCGCAACTTCTG |
| EjSSR15 | LG06_27714710_27714930 | ATTGGGAAGCCGTAGTTGGTGA | TCGCAGCTCTTTCTTCTCACTT |
| EjSSR16 | LG01_38250590_38250807 | AGTTGAGGCTTAGAAGGCAAGG | GAAAGAACTTTGAAAATTCCATCA |
| EjSSR17 | LG12_5320901_5321127 | GAGTGGGACTTCGCATTTGATA | GGAATAACTAACGCCCGCAAC |
| EjSSR18 | LG03_45350948_45351163 | CTCGTTATTGCGTTCAAGCATATT | TTTCTGTTTGGAGCTGGGATTT |
| EjSSR19 | LG16_264901_265127 | CCTTGCCCTGTAGTATTATGTGAT | ATCTGCGGATAAGGAGCACAAG |
| EjSSR20 | LG14_23903168_23903387 | TTTGCGTGATGATCGTTGTCCT | ACACTAAACGGGAGGATGTGGG |
| 品种 Cultivar | 过滤后 reads数 Clean_reads No. | Q30/% | 重测序数 据登录号 Accession number in NGDC | 测序深度 Sequencing depth | SSR序列匹 配reads数 Mapped reads of SSR sequences | SSR序列平均 匹配reads数 Average number of mapped reads for each SSR sequence | 检测到的 位点数 Detected loci |
|---|---|---|---|---|---|---|---|
| 早钟6号 Zaozhong 6 | 32 739 024 | 92.35 | CRR144065 | 12.9× | 3 979 224 | 38.4 | 82 844 |
| 长崎早生 Changqizaosheng | 35 296 384 | 92.02 | CRR144066 | 13.9× | 4 311 146 | 41.6 | 84 013 |
| 茂木 Maomu | 37 259 455 | 92.30 | CRR144067 | 14.7× | 4 439 618 | 42.9 | 84 265 |
| 大五星 Dawuxing | 53 101 602 | 90.54 | CRR144068 | 20.9× | 6 407 584 | 61.9 | 86 560 |
| 月光 Yueguang | 53 088 745 | 91.36 | CRR144069 | 20.9× | 6 406 910 | 61.9 | 86 712 |
| 宁海白 Ninghaibai | 34 771 714 | 91.50 | CRR144070 | 13.7× | 4 207 108 | 40.6 | 86 373 |
| 白玉 Baiyu | 49 199 883 | 90.30 | CRR144071 | 19.4× | 5 915 743 | 57.2 | 86 267 |
| 二八东 Erbadong | 37 032 139 | 92.03 | CRR144072 | 14.6× | 4 016 303 | 38.8 | 84 313 |
| 七星* Sevenstar* | 158 575 610 | 93.41 | CRR078405 | 62.6× | 18 755 558 | 181.2 | 93 943 |
| 火炬* Huoju* | 206 173 911 | 93.16 | CRR056810 | 81.3× | 25 660 354 | 247.9 | 90 345 |
Table 2 Reads of 103 509 SSR sequences mapped to the re-sequencing data of ten loquat samples and detected loci
| 品种 Cultivar | 过滤后 reads数 Clean_reads No. | Q30/% | 重测序数 据登录号 Accession number in NGDC | 测序深度 Sequencing depth | SSR序列匹 配reads数 Mapped reads of SSR sequences | SSR序列平均 匹配reads数 Average number of mapped reads for each SSR sequence | 检测到的 位点数 Detected loci |
|---|---|---|---|---|---|---|---|
| 早钟6号 Zaozhong 6 | 32 739 024 | 92.35 | CRR144065 | 12.9× | 3 979 224 | 38.4 | 82 844 |
| 长崎早生 Changqizaosheng | 35 296 384 | 92.02 | CRR144066 | 13.9× | 4 311 146 | 41.6 | 84 013 |
| 茂木 Maomu | 37 259 455 | 92.30 | CRR144067 | 14.7× | 4 439 618 | 42.9 | 84 265 |
| 大五星 Dawuxing | 53 101 602 | 90.54 | CRR144068 | 20.9× | 6 407 584 | 61.9 | 86 560 |
| 月光 Yueguang | 53 088 745 | 91.36 | CRR144069 | 20.9× | 6 406 910 | 61.9 | 86 712 |
| 宁海白 Ninghaibai | 34 771 714 | 91.50 | CRR144070 | 13.7× | 4 207 108 | 40.6 | 86 373 |
| 白玉 Baiyu | 49 199 883 | 90.30 | CRR144071 | 19.4× | 5 915 743 | 57.2 | 86 267 |
| 二八东 Erbadong | 37 032 139 | 92.03 | CRR144072 | 14.6× | 4 016 303 | 38.8 | 84 313 |
| 七星* Sevenstar* | 158 575 610 | 93.41 | CRR078405 | 62.6× | 18 755 558 | 181.2 | 93 943 |
| 火炬* Huoju* | 206 173 911 | 93.16 | CRR056810 | 81.3× | 25 660 354 | 247.9 | 90 345 |
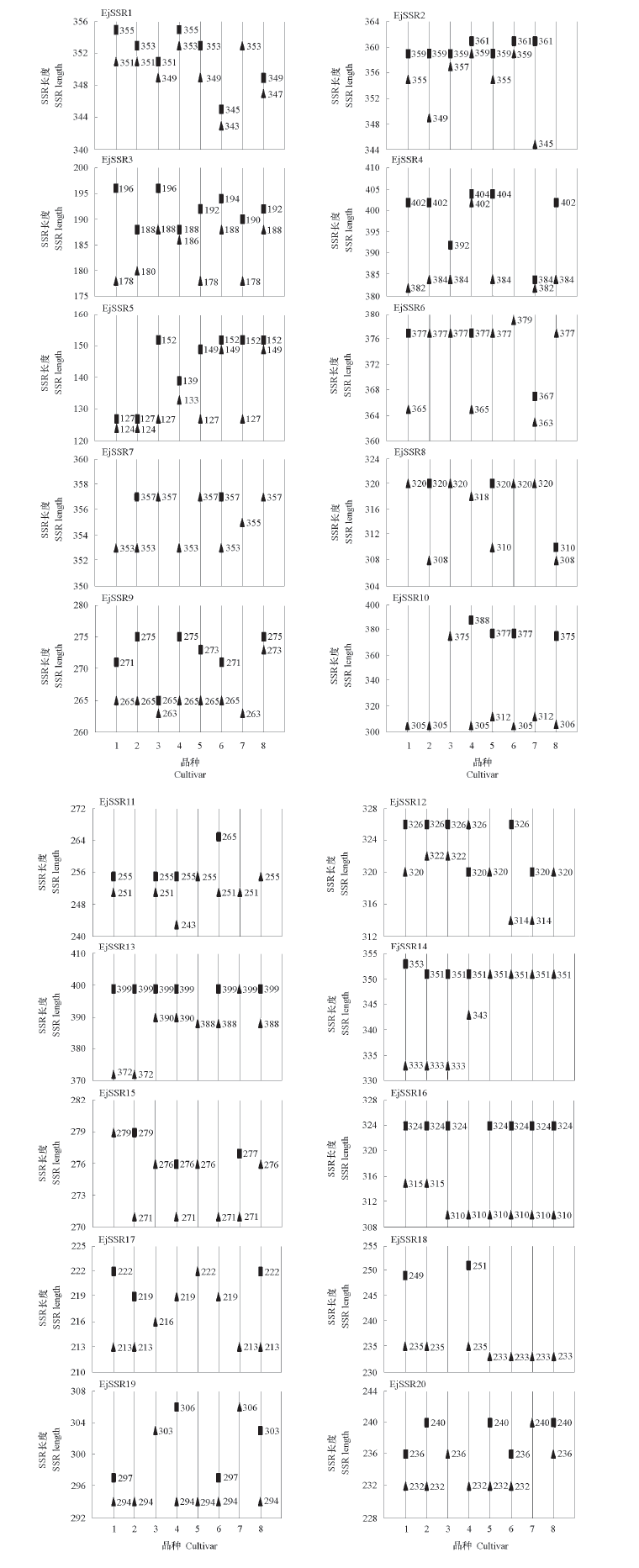
Fig. 5 Fluorescence analysis of the length of PCR product in 20 pairs of primers in 8 cultivars 1. Zaozhong 6;2. Changqizaosheng;3. Maomu;4. Dawuxing;5. Yueguang;6. Ninghaibai;7. Baiyu;8. Erbadong. ▲ and ■ indicate alleles.
| [1] | Ai Ye, Chen Lu, Xie Taixiang, Chen Juan, Lan Siren, Peng Donghui. 2019. Construction of core collection of Cymbidium ensifoliumcultivars based on SSR fluorescent markers . Acta Horticulturae Sinica, 46(10):1999-2008. (in Chinese) |
| 艾叶, 陈璐, 谢泰祥, 陈娟, 兰思仁, 彭东辉. 2019. 基于SSR荧光标记构建建兰品种核心种质. 园艺学报, 46(10):1999-2008. | |
| [2] | Fukuda S, Ishimoto K, Sato S, Terakami S, Hiehata N, Yamamoto T. 2016a. A high-density genetic linkage map of bronze loquat based on SSR and RAPD markers. Tree Genetics & Genomes, 12(4):79-92. |
| [3] |
Fukuda S, Ishimoto K, Terakami S, Yamamoto T, Hiehata N. 2016b. Genetic mapping of the loquat canker resistance gene pse-cin loquat (Eriobotrya japonica). Scientia Horticulturae, 200:19-24.
doi: 10.1016/j.scienta.2015.12.036 URL |
| [4] |
Fukuda S, Nishitani C, Hiehata N, Tominaga Y, Yamamoto T. 2013. Genetic diversity of loquat accessions in Japan as assessed by SSR markers. Journal of the Japanese Society for Horticultural Science, 82(2):131-137.
doi: 10.2503/jjshs1.82.131 URL |
| [5] | Gao Yuan, Wang Kun, Wang Dajiang, Liu Lijun, Li Lianwen, Piao Jicheng. 2019. Genetic diversity and genetic structure of Malus baccata and Malus prunifolia from China as revealed by fluorescent SSR markers . Acta Horticulturae Sinica, 46(7):1225-1237. (in Chinese) |
| 高源, 王昆, 王大江, 刘立军, 李连文, 朴继成. 2019. 中国山荆子和楸子种质资源遗传多样性和遗传结构的荧光SSR分析. 园艺学报, 46(7):1225-1237. | |
| [6] | Gou Wei. 2016. The development and utilization of loquat EST-SSR markers based on transcriptome sequencing[Ph. D. Dissertation]. Chengdu:Sichuan Agricultural University. (in Chinese) |
| 芶维. 2016. 基于转录组测序枇杷EST-SSR标记的开发与利用[博士论文]. 成都:四川农业大学. | |
| [7] | Guan Ling, Zhang Zhen, Wang Xin-wei, Xue Hua-bo, Liu Yan-hong, Wang San-hong, Qiao Yu-shan. 2011. Evaluation and application of the SSR loci in apple genome. China Agriculture Science, 44(21):4415-4428. (in Chinese) |
| 关玲, 章镇, 王新卫, 薛华柏, 刘艳红, 王三红, 乔玉山. 2011. 苹果基因组SSR位点分析与应用. 中国农业科学, 44(21):4415-4428. | |
| [8] | Guo Jun, Zhu Jie, Xie Shangqian, Zhang Ye, Ye Beilei, Zheng Liyan, Ling Peng. 2020. Development of SSR molecular markers based on transcriptome and analysis of genetic relationship of germplasm resources in avocado. Acta Horticulturae Sinica, 47(8):1552-1564. (in Chinese) |
| 郭俊, 朱婕, 谢尚潜, 张叶, 叶蓓蕾, 郑丽燕, 凌鹏. 2020. 油梨转录组SSR分子标记开发与种质资源亲缘关系分析. 园艺学报, 47(8):1552-1564. | |
| [9] |
Gupta P, Varshney R. 2000. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica, 113(3):163-185.
doi: 10.1023/A:1003910819967 URL |
| [10] |
He Q, Li X W, Liang G L, Ji K, Guo Q G, Yuan W M, Zhou G Z, Chen K S, van de Weg W E, Gao Z S. 2011. Genetic diversity and identity of Chinese loquat cultivars/accessions(Eriobotrya japonica)using apple SSR markers. Plant Molecular Biology Reporter, 29(1):197-208.
doi: 10.1007/s11105-010-0218-9 URL |
| [11] |
Jiang S, An H, Xu F, Zhang X. 2020. Chromosome-level genome assembly and annotation of the loquat(Eriobotrya japonica) genome. GigaScience, 9(3):giaa015.
doi: 10.1093/gigascience/giaa015 URL |
| [12] | Jiang Shuang, Luo Jun, Wang Xiao-qing, Shi Chun-hui. 2019. A study on screening efficiency of the polymorphism of SSR markers based on the re-sequencing data in Pyrus . Journal of Fruit Science, 36(2):265-274. (in Chinese) |
| 蒋爽, 骆军, 王晓庆, 施春晖. 2019. 基于基因组重测序数据高效筛选梨SSR标记多态性引物. 果树学报, 36(2):265-274. | |
| [13] |
Kalinowski S T, Taper M L, Marshall T C. 2007. Revising how the computer program cervus,accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16(5):1099-1106.
doi: 10.1111/j.1365-294X.2007.03089.x URL |
| [14] | Li Yongtan, Zhang Jun, Huang Yali, Fan Jianmin, Zhang Yiwen, Zuo Lihui. 2020. Analysis of chloroplast genome ofPyrus betulaefolia . Acta Horticulturae Sinica, 2020, 47(6):1021-1032. (in Chinese) |
| 李泳潭, 张军, 黄亚丽, 范建敏, 张益文, 左力辉. 2020. 杜梨叶绿体基因组分析. 园艺学报, 47(6):1021-1032. | |
| [15] | Liu Shuo, Liu Ning, Zhang Qiuping, Zhang Yuping, Zhang Yujun, Xu Ming, Ma Xiaoxue, Liu Weisheng. 2019. Genetic diversity of the apricot germplasms from North & Northeast China. Acta Horticulturae Sinica, 46(6):1045-1056. (in Chinese) |
| 刘硕, 刘宁, 章秋平, 张玉萍, 张玉君, 徐铭, 马小雪, 刘威生. 2019. 中国华北和东北地区杏种质资源遗传多样性分析. 园艺学报, 46(6):1045-1056. | |
| [16] |
Ouni R, Zborowska A, Sehic J, Choulak S, Hormazae J L, Garkava-Gustavsson L, Mars M. 2020. Genetic diversity and structure of Tunisian local pear germplasm as revealed by SSR markers. Horticultural Plant Journal, 6(2):61-70.
doi: 10.1016/j.hpj.2020.03.003 URL |
| [17] |
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A. 1996. The comparison of RFLP,RAPD,AFLP and SSR(microsatellite)markers for germplasm analysis. Molecular Breeding, 2(3):225-238.
doi: 10.1007/BF00564200 URL |
| [18] |
Soriano J M, Romero C, Vilanova S, Llacer G, Badenes M L. 2005. Genetic diversity of loquat germplasm[Eriobotrya japonica (Thunb) Lindl] assessed by SSR markers. Genome, 48(1):108-114.
pmid: 15729402 |
| [19] | Sun Jun, Li Xiao-ying, Xu Hong-xia, Zhang Lin, Chen Jun-wei. 2018. Identification of white flesh loquat germplasms of Zhejiang province with MCID strategy using genic-SSR markers. Journal of Fruit Science, 35(5):539-547. (in Chinese) |
| 孙钧, 李晓颖, 徐红霞, 张林, 陈俊伟. 2018. 基于genic-SSR标记的MCID法鉴定浙江白沙枇杷地方种质资源. 果树学报, 35(5):539-547. | |
| [20] |
Toth G, Gaspari Z, Jurka J. 2000. Microsatellites in different eukaryotic genomes:survey and analysis. Genome Research, 10(7):967-981.
doi: 10.1101/gr.10.7.967 URL |
| [21] |
Varshney R K, Graner A, Sorrells M E. 2005. Genic microsatellite markers in plants:features and applications. Trends in Biotechnology, 23(1):48-55.
doi: 10.1016/j.tibtech.2004.11.005 URL |
| [22] |
Wen G, Dang J B, Xie Z Y, Wang J Y, Jiang P F, Guo Q G, Liang G L. 2020. Molecular karyotypes of loquat(Eriobotrya japonica)aneuploids can be detected by using SSR markers combined with quantitative PCR irrespective of heterozygosity. Plant Methods, 16(1):22.
doi: 10.1186/s13007-020-00568-7 URL |
| [23] | Xiang Y, Huang C H, Hu Y, Wen J, Li S, Yi T, Chen H, Xiang J, Ma H. 2017. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Molecular Biology Evolution, 34(2):262-281. |
| [24] | Xiong Yan, Zhang Jinzhu, Dong Jie, Wang Li, Gao Peng, Jiang Wan, Che Daidi. 2020. A review of reduced-representation genome sequencing technique and its applications in ornamental plants. Acta Horticulturae Sinica, 47(6):1194-1202. (in Chinese) |
| 熊燕, 张金柱, 董婕, 王力, 高鹏, 姜琬, 车代弟. 2020. 简化基因组测序技术在观赏植物中的应用研究进展. 园艺学报, 47(6):1194-1202. | |
| [25] |
Xue H, Zhang P, Shi T, Yang J, Wang L, Wang S, Su Y, Zhang H, Qiao Y, Li X. 2018. Genome-wide characterization of simple sequence repeats in Pyrus bretschneideri and their application in an analysis of genetic diversity in pear. BMC Genomics, 19(1):473.
doi: 10.1186/s12864-018-4822-7 URL |
| [26] | Zheng Ting-ting, Wei Wei-lin, Yang Xiang-hui, Lin Shun-quan. 2015. Development of SSR molecular markers based on transcriptome sequencing of Eriobotrya japonica . Subtropical Plant Science, 44(4):274-278. (in Chinese) |
| 郑婷婷, 魏伟淋, 杨向晖, 林顺权. 2015. 基于枇杷转录组序列的SSR分子标记引物开发. 亚热带植物科学, 44(4):274-278. |
| [1] | LUO Hailin, YUAN Lei, WENG Hua, YAN Jiahui, GUO Qingyun, WANG Wenqing, MA Xinming. Identification and Analysis of Complete Genomic Sequence of Broad Bean Wilt Virus 2 Pepper Isolate in Qinghai Province [J]. Acta Horticulturae Sinica, 2023, 50(1): 161-169. |
| [2] | ZHANG Zhanwei, RUAN Xiancong, LIN Shunquan, CHEN Jian, FENG Ruixiang, YANG Xianghui, and XU Shejin. A New Early Maturing White-flesh Loquat Cultivar‘Zaojia 8’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 45-46. |
| [3] | SONG Haiyan, SUN Shuxia, LI Jing, CHEN Dong, TU Meiyan, WANG Lingli, XU Zihong, YIN Denggui, and JIANG Guoliang. A New Mid-maturing Loquat Cultivar‘Xishu 3’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 47-48. |
| [4] | YANG Xianghui, ZHANG Zhanwei, CHEN Jian, FENG Ruixiang, RUAN Xiancong, XU Shejin, ZHAO Chongbin, LI Shuqing, CHEN Jun, and LIN Shunquan, . A New White-flesh Loquat Cultivar‘Yuehui’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 49-50. |
| [5] | PAN Haifa, LU Weiming, GAO Xia, SHENG Yu, ZHOU Hui, XIE Qingmei, SHI Pei, WANG Jiabin, SUN Xiaoyao, and ZHANG Jinyun, . A New Late Maturing Loquat Cultivar‘Huihong 2’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 51-52. |
| [6] | PAN Haifa, LU Weiming, SHENG Yu, ZHOU Hui, XIE Qingmei, CHEN Hongli, WANG Jiabin, ZHANG Xiuying, SHI Pei, and ZHANG Jinyun, . A New Early Maturing Loquat Cultivar‘Dayemen’ [J]. Acta Horticulturae Sinica, 2022, 49(S1): 53-54. |
| [7] | ZHOU Chenping, YANG Min, GUO Jinju, KUANG Ruibin, YANG Hu, HUANG Bingxiong, WEI Yuerong. Dynamic Changes in DNA Methylome and Transcriptome Patterns During Papaya Fruit Ripening [J]. Acta Horticulturae Sinica, 2022, 49(3): 519-532. |
| [8] | WU Junyue, SUN Xueyan, YANG Zhenhua, LUO Lu, MO Cuiyuan, MA Aimin. Methylation Analysis of Mycelium and Sclerotium Genome in Pleurotus tuber-regium [J]. Acta Horticulturae Sinica, 2022, 49(1): 148-156. |
| [9] | YANG Zhiwu, DENG Qunxian, WANG Yongqing, DU Kui, PAN Cuiping, ZHANG Hui, ZHANG Huifen, and WANG Yang. A New Spring Flowering Loquat Cultivar‘Chunhua 1’ [J]. Acta Horticulturae Sinica, 2021, 48(S2): 2819-2820. |
| [10] | JIA Huixia, LI Xixiang, SONG Jiangping, LIN Yu’e, ZHANG Xiaohui, QIU Yang, YANG Wenlong, LOU Qunfeng, WANG Haiping. Genome-Wide Association Study of Powdery Mildew Resistance of Cucumber Core Germplasms [J]. Acta Horticulturae Sinica, 2021, 48(7): 1371-1385. |
| [11] | JIANG Xibing, ZHANG Pingsheng, XU Yang, WU Conglian, ZHANG Dongbei, GONG Bangchu, WU Kaiyun, LAI Junsheng. Genetic Diversity of F1 Hybrids of Chestnut Based on SSR Markers [J]. Acta Horticulturae Sinica, 2021, 48(5): 897-907. |
| [12] | DONG Yi, FENG Yufei, XU Zhongmin, WANG Shimin, TANG Honglü, HUANG Wei. Analysis of the Relationship Between Genetic Distance and Heterosis by SSR Markers in Cabbage(Brassica oleracea var. capitata) [J]. Acta Horticulturae Sinica, 2021, 48(5): 934-946. |
| [13] | HUANG Weijian, LI Meng. Status and Prospects Whole Genome Sequencing in Fruit Trees [J]. Acta Horticulturae Sinica, 2021, 48(4): 733-748. |
| [14] | CHEN Li, XUE Liangjiao, LI Shuxian. Genome-wide Association Study of Flower Color Trait in Prunus persicaf.versicolor [J]. Acta Horticulturae Sinica, 2021, 48(3): 553-565. |
| [15] | LIU Mingxiu, WANG Hongding, ZHANG Yuna, ZHANG Bangyan, LIU Song, LI Yunjia, DANG Jiangbo, HE Qiao, LIANG Guolu, GUO Qigao. Comparative Analysis of the Photosystem Activity of Triploid Loquats and Their Tetraploid,Diploid Parents [J]. Acta Horticulturae Sinica, 2021, 48(1): 37-48. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd