
Acta Horticulturae Sinica ›› 2021, Vol. 48 ›› Issue (9): 1755-1767.doi: 10.16420/j.issn.0513-353x.2020-0953
• Research Papers • Previous Articles Next Articles
WANG Xin, LI Mingyang, TIAN Lin, LIU Dongyun*( )
)
Received:2021-01-18
Revised:2021-05-19
Online:2021-09-25
Published:2021-09-30
Contact:
LIU Dongyun
E-mail:Liudongyun0505@163.com
CLC Number:
WANG Xin, LI Mingyang, TIAN Lin, LIU Dongyun. ISSR and rDNA-ITS Sequence Analysis of the Genetic Relationship of Clematis in Hebei Province[J]. Acta Horticulturae Sinica, 2021, 48(9): 1755-1767.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.ahs.ac.cn/EN/10.16420/j.issn.0513-353x.2020-0953
| 编号 No. | 种 Species | 植株类型 Plant type | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 生活型 Life form | 叶型 Leaf form | 花萼 Calyx | 花色 Sepals color | 花型 Flower form | 花期 (月份) Blooming period | 果期 (月份) Fruiting period | 采集地 Collection place | ||
| 1 | 黄花铁线莲 Clematis intricata | 木质藤本 Woody vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 张家口市蔚县 Yu County,Zhangjiakou City |
| 2 | 芹叶铁线莲 C. aethusifolia | 草质藤本 Herbaceous vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 淡黄色 Canary yellow | 钟状 Bell shape | 7 ~ 8 | 9 | 邯郸市武安市 Wu’an City,Handan City |
| 3 | 褐毛铁线莲 C. fusca | 草质藤本 Herbaceous vines | 一回羽状复叶 One pinnately compound leaves | 4 | 紫色 Purple | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 4 | 钝萼铁线莲 C. peterae | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 ~ 5 | 白色 White | 轮状 Wheel shape | 5 ~ 6 | 7 ~ 8 | 邯郸市磁县 Ci County,Handan City |
| 5 | 大叶铁线莲 C. heracleifolia | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 9 ~ 10 | 保定市易县 Yi County,Baoding City |
| 6 | 毛果扬子铁线莲 C. puberula | 草质藤本 Herbaceous vines | 一回羽状或二回三出复叶One pinnately or two ternately compound leaves | 4 | 白色 White | 十字型 Cross | 7 ~ 9 | 9 ~ 10 | 邯郸市磁县 Ci County,Handan City |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市满城区 Mancheng District, Baoding City |
| 8 | 太行铁线莲 C. kirilowii | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市易县 Yi County,Baoding City |
| 9 | 短尾铁线莲 C. brevicaudata | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 7 ~ 8 | 9 ~ 10 | 保定市阜平县 Fuping County,Baoding City |
| 10 | 卷萼铁线莲 C. tubulosa | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 10 | 承德市兴隆县 Xinglong County, Chengde City |
| 11 | 粗齿铁线莲 C. argentilucida | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 5 ~ 7 | 7 ~ 10 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 12 | 灌木铁线莲 C. fruticosa | 灌木 Shrub | 一回羽状复叶 One pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 7 ~ 8 | 10 | 张家口市涿鹿县 Zhuolu County, Zhangjiakou City |
| 13 | 棉团铁线莲 C. hexapetala | 木质藤本 Woody vines | 单叶 Simple leaf | 4 ~ 8 | 白色 White | 轮状 Wheel shape | 6 ~ 7 | 8 ~ 9 | 承德市兴隆县 Xinglong County, Chengde City |
| 14 | 羽叶铁线莲 C. pinnata | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 筒状 Tubular | 9 ~ 10 | 11 ~ 12 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 15 | 长瓣铁线莲 C. macropetala | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 蓝色或 淡紫色 Blue or lavender | 钟状 Bell shape | 7 | 8 | 保定市易县 Yi County,Baoding City |
| 16 | 槭叶铁线莲 C. acerfolia | 灌木 Shrub | 单叶 Simple leaf | 5 ~ 8 | 白色或 粉红色 White or pink | 高脚 碟状 Salver- form | 3月中旬 ~ 4月中旬 | 5 ~ 6 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 17 | 半钟铁线莲 C. ochotensis | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 淡蓝色 Light blue | 钟状 Bell shape | 5 ~ 6 | 7 ~ 8 | 保定市易县 Yi County,Baoding City |
Table 1 The morphological characteristics and collect information of the tested Clematis L.
| 编号 No. | 种 Species | 植株类型 Plant type | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 生活型 Life form | 叶型 Leaf form | 花萼 Calyx | 花色 Sepals color | 花型 Flower form | 花期 (月份) Blooming period | 果期 (月份) Fruiting period | 采集地 Collection place | ||
| 1 | 黄花铁线莲 Clematis intricata | 木质藤本 Woody vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 张家口市蔚县 Yu County,Zhangjiakou City |
| 2 | 芹叶铁线莲 C. aethusifolia | 草质藤本 Herbaceous vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 淡黄色 Canary yellow | 钟状 Bell shape | 7 ~ 8 | 9 | 邯郸市武安市 Wu’an City,Handan City |
| 3 | 褐毛铁线莲 C. fusca | 草质藤本 Herbaceous vines | 一回羽状复叶 One pinnately compound leaves | 4 | 紫色 Purple | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 4 | 钝萼铁线莲 C. peterae | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 ~ 5 | 白色 White | 轮状 Wheel shape | 5 ~ 6 | 7 ~ 8 | 邯郸市磁县 Ci County,Handan City |
| 5 | 大叶铁线莲 C. heracleifolia | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 9 ~ 10 | 保定市易县 Yi County,Baoding City |
| 6 | 毛果扬子铁线莲 C. puberula | 草质藤本 Herbaceous vines | 一回羽状或二回三出复叶One pinnately or two ternately compound leaves | 4 | 白色 White | 十字型 Cross | 7 ~ 9 | 9 ~ 10 | 邯郸市磁县 Ci County,Handan City |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市满城区 Mancheng District, Baoding City |
| 8 | 太行铁线莲 C. kirilowii | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市易县 Yi County,Baoding City |
| 9 | 短尾铁线莲 C. brevicaudata | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 7 ~ 8 | 9 ~ 10 | 保定市阜平县 Fuping County,Baoding City |
| 10 | 卷萼铁线莲 C. tubulosa | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 10 | 承德市兴隆县 Xinglong County, Chengde City |
| 11 | 粗齿铁线莲 C. argentilucida | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 5 ~ 7 | 7 ~ 10 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 12 | 灌木铁线莲 C. fruticosa | 灌木 Shrub | 一回羽状复叶 One pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 7 ~ 8 | 10 | 张家口市涿鹿县 Zhuolu County, Zhangjiakou City |
| 13 | 棉团铁线莲 C. hexapetala | 木质藤本 Woody vines | 单叶 Simple leaf | 4 ~ 8 | 白色 White | 轮状 Wheel shape | 6 ~ 7 | 8 ~ 9 | 承德市兴隆县 Xinglong County, Chengde City |
| 14 | 羽叶铁线莲 C. pinnata | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 筒状 Tubular | 9 ~ 10 | 11 ~ 12 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 15 | 长瓣铁线莲 C. macropetala | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 蓝色或 淡紫色 Blue or lavender | 钟状 Bell shape | 7 | 8 | 保定市易县 Yi County,Baoding City |
| 16 | 槭叶铁线莲 C. acerfolia | 灌木 Shrub | 单叶 Simple leaf | 5 ~ 8 | 白色或 粉红色 White or pink | 高脚 碟状 Salver- form | 3月中旬 ~ 4月中旬 | 5 ~ 6 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 17 | 半钟铁线莲 C. ochotensis | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 淡蓝色 Light blue | 钟状 Bell shape | 5 ~ 6 | 7 ~ 8 | 保定市易县 Yi County,Baoding City |
| 引物名称 Primer name | 引物序列 Primer sequence | 退火温度/℃ Annealing temperature | 位点数 Number of loci | 多态性位点数 Number of polymorphic loci | 多态性位点百分率/% Percentage of polymorphic loci |
|---|---|---|---|---|---|
| U815 | CTCTCTCTCTCTCTCTG | 54.0 | 11 | 11 | 100 |
| U824 | TCTCTCTCTCTCTCTCG | 55.4 | 12 | 12 | 100 |
| U834 | AGAGAGAGAGAGAGAGYT | 57.5 | 7 | 7 | 100 |
| U835 | AGAGAGAGAGAGAGAGYC | 50.4 | 9 | 9 | 100 |
| U836 | AGAGAGAGAGAGAGAGYA | 55.4 | 6 | 6 | 100 |
| U840 | GAGAGAGAGAGAGAGAYT | 55.4 | 7 | 7 | 100 |
| U841 | GAGAGAGAGAGAGAGAYC | 55.4 | 10 | 10 | 100 |
| U843 | CTCTCTCTCTCTCTCTRA | 54.0 | 8 | 8 | 100 |
| U844 | CTCTCTCTCTCTCTCTRC | 55.4 | 6 | 6 | 100 |
| U845 | CTCTCTCTCTCTCTCTRG | 55.4 | 10 | 10 | 100 |
| U866 | CTC CTC CTC CTC CTC CTC | 55.4 | 7 | 7 | 100 |
| U899 | CATGGTGTTGGTCATTGTTCCA | 51.8 | 11 | 11 | 100 |
Table 2 Primers information and polymorphism used for ISSR-PCR reaction of genomic DNA from 17 plants of Clematis L.
| 引物名称 Primer name | 引物序列 Primer sequence | 退火温度/℃ Annealing temperature | 位点数 Number of loci | 多态性位点数 Number of polymorphic loci | 多态性位点百分率/% Percentage of polymorphic loci |
|---|---|---|---|---|---|
| U815 | CTCTCTCTCTCTCTCTG | 54.0 | 11 | 11 | 100 |
| U824 | TCTCTCTCTCTCTCTCG | 55.4 | 12 | 12 | 100 |
| U834 | AGAGAGAGAGAGAGAGYT | 57.5 | 7 | 7 | 100 |
| U835 | AGAGAGAGAGAGAGAGYC | 50.4 | 9 | 9 | 100 |
| U836 | AGAGAGAGAGAGAGAGYA | 55.4 | 6 | 6 | 100 |
| U840 | GAGAGAGAGAGAGAGAYT | 55.4 | 7 | 7 | 100 |
| U841 | GAGAGAGAGAGAGAGAYC | 55.4 | 10 | 10 | 100 |
| U843 | CTCTCTCTCTCTCTCTRA | 54.0 | 8 | 8 | 100 |
| U844 | CTCTCTCTCTCTCTCTRC | 55.4 | 6 | 6 | 100 |
| U845 | CTCTCTCTCTCTCTCTRG | 55.4 | 10 | 10 | 100 |
| U866 | CTC CTC CTC CTC CTC CTC | 55.4 | 7 | 7 | 100 |
| U899 | CATGGTGTTGGTCATTGTTCCA | 51.8 | 11 | 11 | 100 |
| 编号 No. | 样本 Sample | ITS | ITS1 | ITS2 | |||
|---|---|---|---|---|---|---|---|
| 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | ||
| 1 | 黄花铁线莲 C. intricata | 550 | 60.00 | 164 | 59.15 | 227 | 65.60 |
| 2 | 芹叶铁线莲 C. aethusifolia | 549 | 60.88 | 171 | 60.36 | 219 | 67.10 |
| 3 | 褐毛铁线莲 C. fusca | 561 | 61.50 | 180 | 61.67 | 223 | 66.80 |
| 4 | 钝萼铁线莲 C. peterae | 550 | 61.27 | 172 | 60.47 | 219 | 67.60 |
| 5 | 大叶铁线莲 C. heracleifolia | 550 | 61.64 | 172 | 61.05 | 219 | 68.00 |
| 6 | 毛果扬子铁线莲 C. puberula | 543 | 60.89 | 164 | 58.90 | 220 | 67.70 |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 544 | 61.76 | 165 | 59.39 | 220 | 70.00 |
| 8 | 太行铁线莲 C. kirilowii | 545 | 61.76 | 165 | 59.76 | 221 | 69.70 |
| 9 | 短尾铁线莲 C. brevicaudata | 550 | 61.38 | 172 | 60.47 | 219 | 67.90 |
| 10 | 卷萼铁线莲 C. tubulosa | 550 | 61.57 | 172 | 61.05 | 219 | 67.90 |
| 11 | 粗齿铁线莲 C. argentilucida | 550 | 61.68 | 172 | 60.59 | 219 | 68.50 |
| 12 | 灌木铁线莲 C. fruticosa | 543 | 60.04 | 165 | 56.97 | 219 | 67.10 |
| 13 | 棉团铁线莲 C. hexapetala | 549 | 61.38 | 169 | 59.17 | 221 | 69.20 |
| 14 | 羽叶铁线莲 C. pinnata | 550 | 61.79 | 172 | 61.05 | 219 | 68.50 |
| 15 | 长瓣铁线莲 C. macropetala | 548 | 60.95 | 171 | 59.65 | 218 | 67.40 |
| 16 | 槭叶铁线莲 C. acerfolia | 559 | 62.79 | 180 | 63.89 | 220 | 68.60 |
| 17 | 半钟铁线莲 C. ochotensis | 557 | 61.40 | 179 | 60.89 | 219 | 67.60 |
Table 3 Length of ITS(including ITSl and ITS2)and content of G + C
| 编号 No. | 样本 Sample | ITS | ITS1 | ITS2 | |||
|---|---|---|---|---|---|---|---|
| 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | ||
| 1 | 黄花铁线莲 C. intricata | 550 | 60.00 | 164 | 59.15 | 227 | 65.60 |
| 2 | 芹叶铁线莲 C. aethusifolia | 549 | 60.88 | 171 | 60.36 | 219 | 67.10 |
| 3 | 褐毛铁线莲 C. fusca | 561 | 61.50 | 180 | 61.67 | 223 | 66.80 |
| 4 | 钝萼铁线莲 C. peterae | 550 | 61.27 | 172 | 60.47 | 219 | 67.60 |
| 5 | 大叶铁线莲 C. heracleifolia | 550 | 61.64 | 172 | 61.05 | 219 | 68.00 |
| 6 | 毛果扬子铁线莲 C. puberula | 543 | 60.89 | 164 | 58.90 | 220 | 67.70 |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 544 | 61.76 | 165 | 59.39 | 220 | 70.00 |
| 8 | 太行铁线莲 C. kirilowii | 545 | 61.76 | 165 | 59.76 | 221 | 69.70 |
| 9 | 短尾铁线莲 C. brevicaudata | 550 | 61.38 | 172 | 60.47 | 219 | 67.90 |
| 10 | 卷萼铁线莲 C. tubulosa | 550 | 61.57 | 172 | 61.05 | 219 | 67.90 |
| 11 | 粗齿铁线莲 C. argentilucida | 550 | 61.68 | 172 | 60.59 | 219 | 68.50 |
| 12 | 灌木铁线莲 C. fruticosa | 543 | 60.04 | 165 | 56.97 | 219 | 67.10 |
| 13 | 棉团铁线莲 C. hexapetala | 549 | 61.38 | 169 | 59.17 | 221 | 69.20 |
| 14 | 羽叶铁线莲 C. pinnata | 550 | 61.79 | 172 | 61.05 | 219 | 68.50 |
| 15 | 长瓣铁线莲 C. macropetala | 548 | 60.95 | 171 | 59.65 | 218 | 67.40 |
| 16 | 槭叶铁线莲 C. acerfolia | 559 | 62.79 | 180 | 63.89 | 220 | 68.60 |
| 17 | 半钟铁线莲 C. ochotensis | 557 | 61.40 | 179 | 60.89 | 219 | 67.60 |
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 29 | 47 | 48 | 49 | 53 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 79 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | T | A | G | C | A | G | G | C | G | — | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | T | A | G | C | G | A | G | C | G | T | C | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | T | A | G | C | A | A | G | C | G | C | C | G | C | C | C | C | G | G | C | G | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | G | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | A | R | C | G | A | G | C | G | C | T | — | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | T | T | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | T | W | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | T | A | G | Y | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | T | A | G | C | A | G | G | A | A | A | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | A | G | C | A | A | C | T | T | A | G | — | C | G | G | — | — | — | — | — | — | — | G | G | A | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | T | A | G | C | G | A | A | C | G | C | G | — | C | C | C | — | — | — | — | — | — | — | C | G | G | T | C | C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | A | G | C | A | G | G | C | G | C | C | G | C | C | T | C | C | — | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | T | A | G | C | G | C | A | C | G | C | G | — | C | C | T | C | C | G | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | C | A | C | A | A | T | T | A | C | C | G | G | T | A | G | A | A | C | T | T | T | A | T | — | C | C | C | T | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | C | T | A | W | T | T | A | C | C | K | G | T | A | G | A | A | T | T | T | T | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | C | T | — | T | G | T | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | T | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | C | T | T | T | A | C | C | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp | Loci | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | C | C | A | A | T | T | A | A | C | R | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | C | C | G | A | T | T | A | A | C | G | G | T | A | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | C | C | A | A | T | T | A | G | A | G | G | T | A | G | G | A | C | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | T | T | A | C | — | G | A | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | C | T | A | A | C | C | G | A | C | G | G | C | G | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | C | T | A | A | T | T | A | A | C | G | G | T | T | G | A | A | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 407 | 408 | 416 | 428 | 429 | 464 | 469 | 470 | 475 | 488 | 492 | 499 | 505 | 532 | 539 | 541 | 544 | 546 | 547 | 548 | 549 | 550 | 551 | 552 | 553 | 554 | 556 | 558 | 559 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | A | G | T | T | T | T | T | T | C | T | C | C | T | G | T | T | T | C | C | C | G | G | T | C | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | C | A | C | A | C | G | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | G | T | T | T | T | T | T | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | G | T | T | T | T | C | C | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | C | A | T | T | T | T | T | C | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | - | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | G | C | T | T | T | T | T | T | G | C | C | T | R | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | G | T | T | T | T | C | C | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | G | T | T | T | C | T | T | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | G | T | T | T | T | C | C | C | G | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | A | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | G | C | T | T | T | Y | Y | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | G | T | T | T | T | T | T | C | C | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | G | T | C | C | T | T | C | C | G | C | T | C | G | T | T | C | G | G | — | — | — | — | — | T | A | A | C | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | G | T | T | T | T | T | T | C | G | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 4 Singletom specific discrimination site of 17 species in Clematis
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 29 | 47 | 48 | 49 | 53 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 79 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | T | A | G | C | A | G | G | C | G | — | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | T | A | G | C | G | A | G | C | G | T | C | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | T | A | G | C | A | A | G | C | G | C | C | G | C | C | C | C | G | G | C | G | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | G | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | A | R | C | G | A | G | C | G | C | T | — | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | T | T | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | T | W | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | T | A | G | Y | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | T | A | G | C | A | G | G | A | A | A | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | A | G | C | A | A | C | T | T | A | G | — | C | G | G | — | — | — | — | — | — | — | G | G | A | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | T | A | G | C | G | A | A | C | G | C | G | — | C | C | C | — | — | — | — | — | — | — | C | G | G | T | C | C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | A | G | C | A | G | G | C | G | C | C | G | C | C | T | C | C | — | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | T | A | G | C | G | C | A | C | G | C | G | — | C | C | T | C | C | G | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | C | A | C | A | A | T | T | A | C | C | G | G | T | A | G | A | A | C | T | T | T | A | T | — | C | C | C | T | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | C | T | A | W | T | T | A | C | C | K | G | T | A | G | A | A | T | T | T | T | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | C | T | — | T | G | T | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | T | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | C | T | T | T | A | C | C | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp | Loci | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | C | C | A | A | T | T | A | A | C | R | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | C | C | G | A | T | T | A | A | C | G | G | T | A | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | C | C | A | A | T | T | A | G | A | G | G | T | A | G | G | A | C | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | T | T | A | C | — | G | A | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | C | T | A | A | C | C | G | A | C | G | G | C | G | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | C | T | A | A | T | T | A | A | C | G | G | T | T | G | A | A | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 407 | 408 | 416 | 428 | 429 | 464 | 469 | 470 | 475 | 488 | 492 | 499 | 505 | 532 | 539 | 541 | 544 | 546 | 547 | 548 | 549 | 550 | 551 | 552 | 553 | 554 | 556 | 558 | 559 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | A | G | T | T | T | T | T | T | C | T | C | C | T | G | T | T | T | C | C | C | G | G | T | C | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | C | A | C | A | C | G | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | G | T | T | T | T | T | T | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | G | T | T | T | T | C | C | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | C | A | T | T | T | T | T | C | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | - | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | G | C | T | T | T | T | T | T | G | C | C | T | R | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | G | T | T | T | T | C | C | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | G | T | T | T | C | T | T | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | G | T | T | T | T | C | C | C | G | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | A | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | G | C | T | T | T | Y | Y | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | G | T | T | T | T | T | T | C | C | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | G | T | C | C | T | T | C | C | G | C | T | C | G | T | T | C | G | G | — | — | — | — | — | T | A | A | C | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | G | T | T | T | T | T | T | C | G | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本编 号No. | 遗传相似距离 Genetic distances among 17 plants in Clematis L. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| 1 | |||||||||||||||||
| 2 | 0.025 | ||||||||||||||||
| 3 | 0.029 | 0.015 | |||||||||||||||
| 4 | 0.035 | 0.021 | 0.031 | ||||||||||||||
| 5 | 0.033 | 0.015 | 0.025 | 0.006 | |||||||||||||
| 6 | 0.037 | 0.023 | 0.033 | 0.013 | 0.008 | ||||||||||||
| 7 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | |||||||||||
| 8 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | 0 | ||||||||||
| 9 | 0.035 | 0.017 | 0.027 | 0.008 | 0.002 | 0.01 | 0.037 | 0.037 | |||||||||
| 10 | 0.033 | 0.015 | 0.025 | 0.006 | 0 | 0.008 | 0.039 | 0.039 | 0.002 | ||||||||
| 11 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.008 | 0.039 | 0.039 | 0.006 | 0.004 | |||||||
| 12 | 0.031 | 0.021 | 0.029 | 0.033 | 0.029 | 0.037 | 0.045 | 0.045 | 0.031 | 0.029 | 0.033 | ||||||
| 13 | 0.041 | 0.035 | 0.045 | 0.043 | 0.039 | 0.039 | 0.023 | 0.023 | 0.041 | 0.039 | 0.039 | 0.039 | |||||
| 14 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.011 | 0.035 | 0.035 | 0.002 | 0.004 | 0.008 | 0.033 | 0.039 | ||||
| 15 | 0.041 | 0.017 | 0.025 | 0.023 | 0.017 | 0.021 | 0.037 | 0.037 | 0.019 | 0.017 | 0.017 | 0.035 | 0.041 | 0.021 | |||
| 16 | 0.049 | 0.031 | 0.039 | 0.049 | 0.043 | 0.051 | 0.057 | 0.057 | 0.045 | 0.043 | 0.047 | 0.033 | 0.055 | 0.047 | 0.045 | ||
| 17 | 0.039 | 0.015 | 0.027 | 0.023 | 0.017 | 0.021 | 0.039 | 0.039 | 0.019 | 0.017 | 0.017 | 0.031 | 0.039 | 0.021 | 0.009 | 0.039 | |
Table 5 Pairwise divergence of ITS sequences using Kimura-2 parameter distance
| 样本编 号No. | 遗传相似距离 Genetic distances among 17 plants in Clematis L. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| 1 | |||||||||||||||||
| 2 | 0.025 | ||||||||||||||||
| 3 | 0.029 | 0.015 | |||||||||||||||
| 4 | 0.035 | 0.021 | 0.031 | ||||||||||||||
| 5 | 0.033 | 0.015 | 0.025 | 0.006 | |||||||||||||
| 6 | 0.037 | 0.023 | 0.033 | 0.013 | 0.008 | ||||||||||||
| 7 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | |||||||||||
| 8 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | 0 | ||||||||||
| 9 | 0.035 | 0.017 | 0.027 | 0.008 | 0.002 | 0.01 | 0.037 | 0.037 | |||||||||
| 10 | 0.033 | 0.015 | 0.025 | 0.006 | 0 | 0.008 | 0.039 | 0.039 | 0.002 | ||||||||
| 11 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.008 | 0.039 | 0.039 | 0.006 | 0.004 | |||||||
| 12 | 0.031 | 0.021 | 0.029 | 0.033 | 0.029 | 0.037 | 0.045 | 0.045 | 0.031 | 0.029 | 0.033 | ||||||
| 13 | 0.041 | 0.035 | 0.045 | 0.043 | 0.039 | 0.039 | 0.023 | 0.023 | 0.041 | 0.039 | 0.039 | 0.039 | |||||
| 14 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.011 | 0.035 | 0.035 | 0.002 | 0.004 | 0.008 | 0.033 | 0.039 | ||||
| 15 | 0.041 | 0.017 | 0.025 | 0.023 | 0.017 | 0.021 | 0.037 | 0.037 | 0.019 | 0.017 | 0.017 | 0.035 | 0.041 | 0.021 | |||
| 16 | 0.049 | 0.031 | 0.039 | 0.049 | 0.043 | 0.051 | 0.057 | 0.057 | 0.045 | 0.043 | 0.047 | 0.033 | 0.055 | 0.047 | 0.045 | ||
| 17 | 0.039 | 0.015 | 0.027 | 0.023 | 0.017 | 0.021 | 0.039 | 0.039 | 0.019 | 0.017 | 0.017 | 0.031 | 0.039 | 0.021 | 0.009 | 0.039 | |
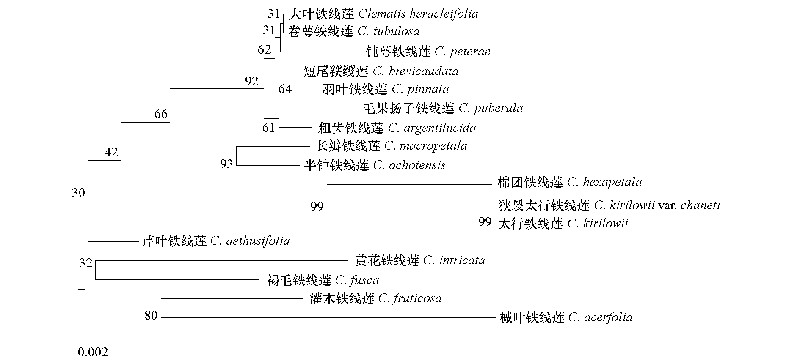
Fig. 5 Molecular phylogenetic tree inferred from ITS of 17 species in Clematis The length of the scale is 0.002,which can be used to calculate the genetic distance of each branch.
| [1] | Jiang Ming, Zhou Ying-qiao, Li Rong-rong. 2011. Sequence analysis of ITS of 8 medicinal plants of Clematis. Chinese Herbal Medicine, 42(9):1802-1806. (in Chinese) |
| 蒋明, 周英巧, 李嵘嵘. 2011. 铁线莲属8种药用植物ITS序列分析. 中草药, 42(9):1802-1806. | |
| [2] | Li Xiao, Wang Pei, Zhao Long, Ju Ai-hua. 2012. rDNA-ITS sequence analysis of Clematis mongolicum. Journal of Inner Mongolia Medical College, 34(4):325-330. (in Chinese) |
| 李骁, 王佩, 赵龙, 鞠爱华. 2012. 铁线莲属蒙药材的rDNA-ITS序列分析. 内蒙古医学院学报, 34(4):325-330. | |
| [3] | Liu Jing-jing, Gao Yi-ke. 2013. Investigation on germplasm resources of clematis in Beijing area. Heilongjiang Agricultural Science,(4):65-69. (in Chinese) |
| 刘晶晶, 高亦珂. 2013. 北京地区野生铁线莲属植物种质资源调查研究. 黑龙江农业科学,(4):65-69. | |
| [4] | Liu Qing-chao, Wang Kui-ling, Lu Wan-pei, Liu Qing-hua. 2014. Reasearch progress of Clematis in China. Journal of Plant Genetic Resource, 15(3):483-490,497. (in Chinese) |
| 刘庆超, 王奎玲, 卢婉佩, 刘庆华. 2014. 我国铁线莲属植物资源研究进展. 植物遗传资源学报, 15(3):483-490,497. | |
| [5] | Mu Lin, Xie Lei. 2011. Systematic location of Clematis acerfolia—analysis of ITS and chloroplast DNA sequences. Journal of Beijing Forestry University, 33(5):49-55. (in Chinese) |
| 穆琳, 谢磊. 2011. 槭叶铁线莲的系统位置初探——来自ITS和叶绿体DNA序列片段的分析. 北京林业大学学报, 33(5):49-55. | |
| [6] | Qian Ren-juan, Zheng Jian, Hu Qing-di, Ma Xiao-hua, Zhang Xu-Le. 2017. Research progress on ornamental germplasm resources of Clematis in China. Chinese Agricultural Science Bulletin, 33(21):75-81. (in Chinese) |
| 钱仁卷, 郑坚, 胡青荻, 马晓华, 张旭乐. 2017. 中国铁线莲属观赏种质资源研究进展. 中国农学通报, 33(21):75-81. | |
| [7] |
Salem S A, Muhammad A K, Hussein M M, Ehab H E, Muhammad A, Muhammad F. 2019. Biochemical and molecular characterization of cowpea landraces using seed storage protein and SRAP marker patterns. Saudi Journal of Biological Sciences, 26(1):74-82.
doi: 10.1016/j.sjbs.2018.09.004 URL |
| [8] | Sheng Lu. 2015. Karyotype analysis and molecular systematics of Clematis[Ph. D. Dissertation]. Nanjing:Nanjing Forestry University. |
| [9] | 盛璐. 2015. 铁线莲属植物的核型分析及分子系统学研究[博士论文]. 南京:南京林业大学. |
| [10] | Shi Ling-yan, Liu Bao-wei, Gu Xin-ying. 2019. Application of biochemical and ISSR molecular markers in identification of Auricularia auricular. Northern Horticulture, 43(9):136-141. (in Chinese) |
| 史灵燕, 刘保卫, 顾新颖. 2019. 生化和ISSR分子标记在黑木耳品种鉴定中的应用, 北方园艺, 43(9):136-141. | |
| [11] | Sun Zheng-hai, Wang Jin, Li Shi-feng, Lin Kai-wen, Hu Yi-chen, Wang Shi-jin. 2012. Establishment and optimization of ISSR-PCR reaction system for Rattan. Journal of Yunnan Agricultural University(Natural Science), 27(5):746-750. (in Chinese) |
| 孙正海, 王锦, 李世峰, 林开文, 胡祎晨, 王仕锦. 2012. 滑叶藤ISSR-PCR反应体系建立及优化. 云南农业大学学报(自然科学), 27(5):746-750. | |
| [12] | Wang Jiang-yong, Qiao Qian, Zhang Jie, Hu Feng-rong. 2020. Research progress of Clematis and application in garden. Journal of Shandong Agricultural University(Natural Science), 51(2):217-221. (in Chinese) |
| 王江勇, 乔谦, 张杰, 胡凤荣. 2020. 铁线莲属植物研究进展与园林应用. 山东农业大学学报(自然科学版), 51(2):217-221. | |
| [13] | Wang Nan, Wang Jin, Li Zong-yan, Yang Kun-mei, He Hui, Xiao Zhen-dong. 2016. ISSR-PCR reaction system optimization and primer screening of Clematis horticulture varieties. Northern Horticulture,(1):80-83. (in Chinese) |
| 王楠, 王锦, 李宗艳, 杨坤梅, 何辉, 肖振东. 2016. 铁线莲园艺品种ISSR-PCR反应体系优化与引物筛选. 北方园艺,(1):80-83. | |
| [14] | Wang W T, Xie L. 2007. A revision of Clematis sect. Tubulosae(Ranunculaceae). Acta Phytotaxonomica Suiica, 45:425-457. |
| [15] | Wang Wen-cai, Li Liang-qian. 2005. A new classification system of Clematis. Acta Botanica Taxa,(5):431-488. (in Chinese) |
| 王文采, 李良千. 2005. 铁线莲属一新分类系统. 植物分类学报,(5):431-488. | |
| [16] | Xie L, Shi J H, Li L Q. 2005. Identity of Clematis tatarinowii and C. pinnata var. ternatifolia(Ranunculaceae). Annales Botanici Fennici, 42:305-308. |
| [17] |
Xie L, Wen J, Li L Q. 2011. Phylogenetic analyses of Clematis(Ranunculaceae)based on sequences of nuclear ribosomal ITS and three plastid regions. Systematic Botany, 36:907-921.
doi: 10.1600/036364411X604921 URL |
| [18] | Yang Jiu-yan, Yang Jie, Yang Ming-bo, Ju Aihua, Qu bi. 2006. ISSR analysis of medicinal plants of the genus Caragana in Ordos Plateau. Chinese Herbal Medicine, 37(10):1562-1566. (in Chinese) |
| 杨九艳, 杨劼, 杨明博, 鞠爱华, 渠弼. 2006. 鄂尔多斯高原锦鸡儿属药用植物的ISSR分析. 中草药, 37(10):1562-1566. | |
| [19] | Zhao Huan, Wu Wei, Zheng You-liang, Pan Hong-mei, Zhai Juan-yuan. 2009. Application of ribosomal DNA ITS sequence analysis in the study of medicinal plants. Shi Zhen Chinese Medicine, 20(4):959-962. (in Chinese) |
| 赵欢, 吴卫, 郑有良, 潘红梅, 翟娟园. 2009. 核糖体DNA ITS序列分析在药用植物研究中的应用. 时珍国医国药, 20(4):959-962. | |
| [20] |
Zhu Hong, Zhu Shu-xia, Li Yong-fu, Yi Xian-gui, Duan Yi-fan, Wang Xian-rong. 2018. Leaf phenotypic variation in natural populations of Cerasus dielsiana. Chinese Journal of Plant Ecology, 42(12):1168-1178. (in Chinese)
doi: 10.17521/cjpe.2018.0196 URL |
| 朱弘, 朱淑霞, 李涌福, 伊贤贵, 段一凡, 王贤荣. 2018. 尾叶樱桃天然种群叶表型性状变异研究. 植物生态学报, 42(12):1168-1178. | |
| [21] | Zhu Yuan-di, Cao Min-ge, Xu Zheng, Wang Kun, Zhang Wen. 2014. Phylogenetic relationship between Xinjiang wild apple(Malus sieversii Roem.) and Chinese apple(Malus × domestica subsp. chinesnsis)based on ITS and matK sequences. Acta Horticulturae Sinica, 41(2):227-239. (in Chinese) |
| 朱元娣, 曹敏格, 许正, 王昆, 张文. 2014. 基于ITS和matK序列探讨新疆野苹果与中国苹果的系统演化关系. 园艺学报, 41(2):227-239. |
| [1] | WU Fan, JIANG Junhao, LU Shan, ZHANG Nan, QIU Shuai, WEI Jianfen, SHEN Baichun. Research Progress on Germplasm Resources and Utilization of Hydrangea in China [J]. Acta Horticulturae Sinica, 2022, 49(9): 2037-2050. |
| [2] | LÜ Zhengxin, HE Yanqun, JIA Dongfeng, HUANG Chunhui, ZHONG Min, LIAO Guanglian, ZHU Yi, YUAN Kaichang, LIU Chuanhao, XU Xiaobiao. Genetic Diversity Analysis of Phenotypic Traits for Kiwifruit Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(7): 1571-1581. |
| [3] | LI Pingping, ZHANG Xiang, LIU Yuting, XIE Zhihe, ZHANG Ruihao, ZHAO Kai, LÜ Junheng, WANG Ziran, WEN Jinfen, ZOU Xuexiao, DENG Minghua. Studies on the Relationship Between Pigment Composition and Fruit Coloration of 63 Peppers [J]. Acta Horticulturae Sinica, 2022, 49(7): 1589-1601. |
| [4] | ZHANG Meng, SHAN Yuying, YANG Yebo, ZHAI Feifei, WANG Zhaoshan, JU Guansheng, SUN Zhenyuan, LI Zhenjian. AFLP Analysis of Genetic Resources of Dendrobium from China [J]. Acta Horticulturae Sinica, 2022, 49(6): 1339-1350. |
| [5] | CAI Zhixiang, YAN Juan, SU Ziwen, XU Ziyuan, ZHANG Minghao, SHEN Zhijun, YANG Jun, MA Ruijuan, YU Mingliang. Evaluation of Main Phenolic Compounds in Different Types of Peach Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(5): 1008-1022. |
| [6] | ZHANG Shicai, LI Yifei, WANG Chunping, YANG Xiaomiao, HUANG Qizhong, HUANG Renzhong. Resistance Identification and Evaluation of Pepper Germplasm to Colletotrichum acutatum [J]. Acta Horticulturae Sinica, 2022, 49(4): 885-892. |
| [7] | XU Wan, LIN Yajun, ZHAO Zhuang, ZHOU Zhuang. Advances in Genetic Resources and Breeding Research of Cymbidium [J]. Acta Horticulturae Sinica, 2022, 49(12): 2722-2742. |
| [8] | WU Zhijun, LI Wei, WANG Xinghua, ZHANG Cheng, JIANG Xun, LI Sheng, LUO Liyong, SUN Kang, ZENG Liang. Genetic Relationship Analysis of the Population Germplasms of Section Thea with Unstable Ovary Locule Number Based on SLAF-seq [J]. Acta Horticulturae Sinica, 2022, 49(11): 2455-2470. |
| [9] | ZHAO Liqun, ZHANG Xiaofei, QIU Yanhong, LIU Hui, ZHANG Jian, YANG Jingjing, ZHANG Haijun, XU Xiulan, WEN Changlong. Studies on Detection of Cucumis Melo Endornavirus Using TaqMan RT-qPCR Method [J]. Acta Horticulturae Sinica, 2022, 49(11): 2471-2478. |
| [10] | SU Jiangshuo, JIA Diwen, WANG Siyue, ZHANG Fei, JIANG Jiafu, CHEN Sumei, FANG Weimin, and CHEN Fadi. Retrospection and Prospect of Chrysanthemum Genetic Breeding for Last Six Decades in China [J]. Acta Horticulturae Sinica, 2022, 49(10): 2143-2162. |
| [11] | CHEN Mingkun, CHEN Lu, SUN Weihong, MA Shanhu, LAN Siren, PENG Donghui, LIU Zhongjian, AI Ye. Genetic Diversity Analysis and Core Collection of Cymbidium ensifolium Germplasm Resources [J]. Acta Horticulturae Sinica, 2022, 49(1): 175-186. |
| [12] | LI Yanhong, NIE Jun, ZHENG Jinrong, TAN Delong, ZHANG Changyuan, SHI Liangliang, XIE Yuming. Genetic Diversity Analysis and Multivariate Evaluation of Cherry Tomato by Phenotypic Traits in South China [J]. Acta Horticulturae Sinica, 2021, 48(9): 1717-1730. |
| [13] | NIE Weijie, WANG Chenhe, MA Wanru, SU Jiangshuo, ZHANG Fei, FANG Weimin, CHEN Sumei, CHEN Fadi, GUAN Zhiyong. Comprehensive Evaluation of Flower Branch Characteristics of Cut Chrysanthemums [J]. Acta Horticulturae Sinica, 2021, 48(6): 1150-1162. |
| [14] | PENG Fangfang, LONG Zhijian, WEI Zhaoxin, LI Xunlan, LUO Youjin, HAN Guohui. Association Analysis of SCoT Markers and Leaf Phenotypic Traits in Cherry Germplasm [J]. Acta Horticulturae Sinica, 2021, 48(2): 325-335. |
| [15] | WANG Zhenjiang, LUO Guoqing, DAI Fanwei, XIAO Gengsheng, LIN Sen, LI Zhiyi, TANG Cuiming. Genetic Diversity of 569 Fruit Mulberry Germplasm Resources Based on Eight Agronomic Traits [J]. Acta Horticulturae Sinica, 2021, 48(12): 2375-2384. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2012 Acta Horticulturae Sinica 京ICP备10030308号-2 国际联网备案号 11010802023439
Tel: 010-82109523 E-Mail: yuanyixuebao@126.com
Support by: Beijing Magtech Co.Ltd