
园艺学报 ›› 2021, Vol. 48 ›› Issue (9): 1755-1767.doi: 10.16420/j.issn.0513-353x.2020-0953
收稿日期:2021-01-18
修回日期:2021-05-19
出版日期:2021-09-25
发布日期:2021-09-30
通讯作者:
刘冬云
E-mail:Liudongyun0505@163.com
基金资助:
WANG Xin, LI Mingyang, TIAN Lin, LIU Dongyun*( )
)
Received:2021-01-18
Revised:2021-05-19
Online:2021-09-25
Published:2021-09-30
Contact:
LIU Dongyun
E-mail:Liudongyun0505@163.com
摘要:
通过ISSR和rDNA-ITS 两种分子标记方法对河北省17个类群的野生铁线莲种间亲缘关系进行了鉴定。利用UPGMA法对ISSR扩增的条带结果进行聚类分析,并用PCR法扩增ITS,通过Clustal X,MEGA等软件比较和分析ITS的序列特征,最后对两种分子标记的结果综合分析。结果表明,利用筛选出的ISSR引物扩增出多态性条带104条,多态性百分比为100%,等位基因数(Na)为2.00,有效等位基因数(Ne)为1.3069,Nei’s基因多样性(H)为0.2196,Shannon’s多样性信息指数(I)为0.3675。聚类结果显示,在遗传距离为0.66处,17种野生铁线莲被分为5类;铁线莲的17个铁线莲样本ITS序列长度为543 ~ 561 bp,含变异位点87个,信息位点37个,特异性鉴别位点50个。其中,长瓣铁线莲与半钟铁线莲、太行铁线莲与狭裂太行铁线莲遗传距离为0,槭叶铁线莲与其他种的遗传距离较大,亲缘关系较远。
中图分类号:
王鑫, 李明阳, 田琳, 刘冬云. 利用ISSR标记及rDNA-ITS序列分析河北省野生铁线莲的亲缘关系[J]. 园艺学报, 2021, 48(9): 1755-1767.
WANG Xin, LI Mingyang, TIAN Lin, LIU Dongyun. ISSR and rDNA-ITS Sequence Analysis of the Genetic Relationship of Clematis in Hebei Province[J]. Acta Horticulturae Sinica, 2021, 48(9): 1755-1767.
| 编号 No. | 种 Species | 植株类型 Plant type | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 生活型 Life form | 叶型 Leaf form | 花萼 Calyx | 花色 Sepals color | 花型 Flower form | 花期 (月份) Blooming period | 果期 (月份) Fruiting period | 采集地 Collection place | ||
| 1 | 黄花铁线莲 Clematis intricata | 木质藤本 Woody vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 张家口市蔚县 Yu County,Zhangjiakou City |
| 2 | 芹叶铁线莲 C. aethusifolia | 草质藤本 Herbaceous vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 淡黄色 Canary yellow | 钟状 Bell shape | 7 ~ 8 | 9 | 邯郸市武安市 Wu’an City,Handan City |
| 3 | 褐毛铁线莲 C. fusca | 草质藤本 Herbaceous vines | 一回羽状复叶 One pinnately compound leaves | 4 | 紫色 Purple | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 4 | 钝萼铁线莲 C. peterae | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 ~ 5 | 白色 White | 轮状 Wheel shape | 5 ~ 6 | 7 ~ 8 | 邯郸市磁县 Ci County,Handan City |
| 5 | 大叶铁线莲 C. heracleifolia | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 9 ~ 10 | 保定市易县 Yi County,Baoding City |
| 6 | 毛果扬子铁线莲 C. puberula | 草质藤本 Herbaceous vines | 一回羽状或二回三出复叶One pinnately or two ternately compound leaves | 4 | 白色 White | 十字型 Cross | 7 ~ 9 | 9 ~ 10 | 邯郸市磁县 Ci County,Handan City |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市满城区 Mancheng District, Baoding City |
| 8 | 太行铁线莲 C. kirilowii | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市易县 Yi County,Baoding City |
| 9 | 短尾铁线莲 C. brevicaudata | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 7 ~ 8 | 9 ~ 10 | 保定市阜平县 Fuping County,Baoding City |
| 10 | 卷萼铁线莲 C. tubulosa | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 10 | 承德市兴隆县 Xinglong County, Chengde City |
| 11 | 粗齿铁线莲 C. argentilucida | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 5 ~ 7 | 7 ~ 10 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 12 | 灌木铁线莲 C. fruticosa | 灌木 Shrub | 一回羽状复叶 One pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 7 ~ 8 | 10 | 张家口市涿鹿县 Zhuolu County, Zhangjiakou City |
| 13 | 棉团铁线莲 C. hexapetala | 木质藤本 Woody vines | 单叶 Simple leaf | 4 ~ 8 | 白色 White | 轮状 Wheel shape | 6 ~ 7 | 8 ~ 9 | 承德市兴隆县 Xinglong County, Chengde City |
| 14 | 羽叶铁线莲 C. pinnata | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 筒状 Tubular | 9 ~ 10 | 11 ~ 12 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 15 | 长瓣铁线莲 C. macropetala | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 蓝色或 淡紫色 Blue or lavender | 钟状 Bell shape | 7 | 8 | 保定市易县 Yi County,Baoding City |
| 16 | 槭叶铁线莲 C. acerfolia | 灌木 Shrub | 单叶 Simple leaf | 5 ~ 8 | 白色或 粉红色 White or pink | 高脚 碟状 Salver- form | 3月中旬 ~ 4月中旬 | 5 ~ 6 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 17 | 半钟铁线莲 C. ochotensis | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 淡蓝色 Light blue | 钟状 Bell shape | 5 ~ 6 | 7 ~ 8 | 保定市易县 Yi County,Baoding City |
表1 供试铁线莲形态特征及采集信息
Table 1 The morphological characteristics and collect information of the tested Clematis L.
| 编号 No. | 种 Species | 植株类型 Plant type | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 生活型 Life form | 叶型 Leaf form | 花萼 Calyx | 花色 Sepals color | 花型 Flower form | 花期 (月份) Blooming period | 果期 (月份) Fruiting period | 采集地 Collection place | ||
| 1 | 黄花铁线莲 Clematis intricata | 木质藤本 Woody vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 张家口市蔚县 Yu County,Zhangjiakou City |
| 2 | 芹叶铁线莲 C. aethusifolia | 草质藤本 Herbaceous vines | 一至二回羽状复叶 One to two pinnately compound leaves | 4 | 淡黄色 Canary yellow | 钟状 Bell shape | 7 ~ 8 | 9 | 邯郸市武安市 Wu’an City,Handan City |
| 3 | 褐毛铁线莲 C. fusca | 草质藤本 Herbaceous vines | 一回羽状复叶 One pinnately compound leaves | 4 | 紫色 Purple | 钟状 Bell shape | 6 ~ 7 | 8 ~ 9 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 4 | 钝萼铁线莲 C. peterae | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 ~ 5 | 白色 White | 轮状 Wheel shape | 5 ~ 6 | 7 ~ 8 | 邯郸市磁县 Ci County,Handan City |
| 5 | 大叶铁线莲 C. heracleifolia | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 9 ~ 10 | 保定市易县 Yi County,Baoding City |
| 6 | 毛果扬子铁线莲 C. puberula | 草质藤本 Herbaceous vines | 一回羽状或二回三出复叶One pinnately or two ternately compound leaves | 4 | 白色 White | 十字型 Cross | 7 ~ 9 | 9 ~ 10 | 邯郸市磁县 Ci County,Handan City |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市满城区 Mancheng District, Baoding City |
| 8 | 太行铁线莲 C. kirilowii | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4或 5 ~ 6 | 白色 White | 轮状 Wheel shape | 6 ~ 8 | 8 ~ 9 | 保定市易县 Yi County,Baoding City |
| 9 | 短尾铁线莲 C. brevicaudata | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 7 ~ 8 | 9 ~ 10 | 保定市阜平县 Fuping County,Baoding City |
| 10 | 卷萼铁线莲 C. tubulosa | 灌木 Shrub | 三出复叶 Ternately compound leaves | 4 | 蓝紫色 Bluish violet | 筒状 Tubular | 7 ~ 8 | 10 | 承德市兴隆县 Xinglong County, Chengde City |
| 11 | 粗齿铁线莲 C. argentilucida | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 轮状 Wheel shape | 5 ~ 7 | 7 ~ 10 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 12 | 灌木铁线莲 C. fruticosa | 灌木 Shrub | 一回羽状复叶 One pinnately compound leaves | 4 | 黄色 Yellow | 钟状 Bell shape | 7 ~ 8 | 10 | 张家口市涿鹿县 Zhuolu County, Zhangjiakou City |
| 13 | 棉团铁线莲 C. hexapetala | 木质藤本 Woody vines | 单叶 Simple leaf | 4 ~ 8 | 白色 White | 轮状 Wheel shape | 6 ~ 7 | 8 ~ 9 | 承德市兴隆县 Xinglong County, Chengde City |
| 14 | 羽叶铁线莲 C. pinnata | 木质藤本 Woody vines | 一回羽状复叶 One pinnately compound leaves | 4 | 白色 White | 筒状 Tubular | 9 ~ 10 | 11 ~ 12 | 秦皇岛市青龙县 Qinglong County, Qinhuangdao City |
| 15 | 长瓣铁线莲 C. macropetala | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 蓝色或 淡紫色 Blue or lavender | 钟状 Bell shape | 7 | 8 | 保定市易县 Yi County,Baoding City |
| 16 | 槭叶铁线莲 C. acerfolia | 灌木 Shrub | 单叶 Simple leaf | 5 ~ 8 | 白色或 粉红色 White or pink | 高脚 碟状 Salver- form | 3月中旬 ~ 4月中旬 | 5 ~ 6 | 石家庄市赞皇县 Zanhuang County, Shijiazhuang City |
| 17 | 半钟铁线莲 C. ochotensis | 木质藤本 Woody vines | 二回三出复叶 Bipinnately and ternately compound leaves | 4 | 淡蓝色 Light blue | 钟状 Bell shape | 5 ~ 6 | 7 ~ 8 | 保定市易县 Yi County,Baoding City |
| 引物名称 Primer name | 引物序列 Primer sequence | 退火温度/℃ Annealing temperature | 位点数 Number of loci | 多态性位点数 Number of polymorphic loci | 多态性位点百分率/% Percentage of polymorphic loci |
|---|---|---|---|---|---|
| U815 | CTCTCTCTCTCTCTCTG | 54.0 | 11 | 11 | 100 |
| U824 | TCTCTCTCTCTCTCTCG | 55.4 | 12 | 12 | 100 |
| U834 | AGAGAGAGAGAGAGAGYT | 57.5 | 7 | 7 | 100 |
| U835 | AGAGAGAGAGAGAGAGYC | 50.4 | 9 | 9 | 100 |
| U836 | AGAGAGAGAGAGAGAGYA | 55.4 | 6 | 6 | 100 |
| U840 | GAGAGAGAGAGAGAGAYT | 55.4 | 7 | 7 | 100 |
| U841 | GAGAGAGAGAGAGAGAYC | 55.4 | 10 | 10 | 100 |
| U843 | CTCTCTCTCTCTCTCTRA | 54.0 | 8 | 8 | 100 |
| U844 | CTCTCTCTCTCTCTCTRC | 55.4 | 6 | 6 | 100 |
| U845 | CTCTCTCTCTCTCTCTRG | 55.4 | 10 | 10 | 100 |
| U866 | CTC CTC CTC CTC CTC CTC | 55.4 | 7 | 7 | 100 |
| U899 | CATGGTGTTGGTCATTGTTCCA | 51.8 | 11 | 11 | 100 |
表2 用于17种铁线莲基因组DNA ISSR-PCR反应的引物信息及多态性分析
Table 2 Primers information and polymorphism used for ISSR-PCR reaction of genomic DNA from 17 plants of Clematis L.
| 引物名称 Primer name | 引物序列 Primer sequence | 退火温度/℃ Annealing temperature | 位点数 Number of loci | 多态性位点数 Number of polymorphic loci | 多态性位点百分率/% Percentage of polymorphic loci |
|---|---|---|---|---|---|
| U815 | CTCTCTCTCTCTCTCTG | 54.0 | 11 | 11 | 100 |
| U824 | TCTCTCTCTCTCTCTCG | 55.4 | 12 | 12 | 100 |
| U834 | AGAGAGAGAGAGAGAGYT | 57.5 | 7 | 7 | 100 |
| U835 | AGAGAGAGAGAGAGAGYC | 50.4 | 9 | 9 | 100 |
| U836 | AGAGAGAGAGAGAGAGYA | 55.4 | 6 | 6 | 100 |
| U840 | GAGAGAGAGAGAGAGAYT | 55.4 | 7 | 7 | 100 |
| U841 | GAGAGAGAGAGAGAGAYC | 55.4 | 10 | 10 | 100 |
| U843 | CTCTCTCTCTCTCTCTRA | 54.0 | 8 | 8 | 100 |
| U844 | CTCTCTCTCTCTCTCTRC | 55.4 | 6 | 6 | 100 |
| U845 | CTCTCTCTCTCTCTCTRG | 55.4 | 10 | 10 | 100 |
| U866 | CTC CTC CTC CTC CTC CTC | 55.4 | 7 | 7 | 100 |
| U899 | CATGGTGTTGGTCATTGTTCCA | 51.8 | 11 | 11 | 100 |
| 编号 No. | 样本 Sample | ITS | ITS1 | ITS2 | |||
|---|---|---|---|---|---|---|---|
| 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | ||
| 1 | 黄花铁线莲 C. intricata | 550 | 60.00 | 164 | 59.15 | 227 | 65.60 |
| 2 | 芹叶铁线莲 C. aethusifolia | 549 | 60.88 | 171 | 60.36 | 219 | 67.10 |
| 3 | 褐毛铁线莲 C. fusca | 561 | 61.50 | 180 | 61.67 | 223 | 66.80 |
| 4 | 钝萼铁线莲 C. peterae | 550 | 61.27 | 172 | 60.47 | 219 | 67.60 |
| 5 | 大叶铁线莲 C. heracleifolia | 550 | 61.64 | 172 | 61.05 | 219 | 68.00 |
| 6 | 毛果扬子铁线莲 C. puberula | 543 | 60.89 | 164 | 58.90 | 220 | 67.70 |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 544 | 61.76 | 165 | 59.39 | 220 | 70.00 |
| 8 | 太行铁线莲 C. kirilowii | 545 | 61.76 | 165 | 59.76 | 221 | 69.70 |
| 9 | 短尾铁线莲 C. brevicaudata | 550 | 61.38 | 172 | 60.47 | 219 | 67.90 |
| 10 | 卷萼铁线莲 C. tubulosa | 550 | 61.57 | 172 | 61.05 | 219 | 67.90 |
| 11 | 粗齿铁线莲 C. argentilucida | 550 | 61.68 | 172 | 60.59 | 219 | 68.50 |
| 12 | 灌木铁线莲 C. fruticosa | 543 | 60.04 | 165 | 56.97 | 219 | 67.10 |
| 13 | 棉团铁线莲 C. hexapetala | 549 | 61.38 | 169 | 59.17 | 221 | 69.20 |
| 14 | 羽叶铁线莲 C. pinnata | 550 | 61.79 | 172 | 61.05 | 219 | 68.50 |
| 15 | 长瓣铁线莲 C. macropetala | 548 | 60.95 | 171 | 59.65 | 218 | 67.40 |
| 16 | 槭叶铁线莲 C. acerfolia | 559 | 62.79 | 180 | 63.89 | 220 | 68.60 |
| 17 | 半钟铁线莲 C. ochotensis | 557 | 61.40 | 179 | 60.89 | 219 | 67.60 |
表3 ITS序列(包括ITS1和ITS2)长度及其G + C含量
Table 3 Length of ITS(including ITSl and ITS2)and content of G + C
| 编号 No. | 样本 Sample | ITS | ITS1 | ITS2 | |||
|---|---|---|---|---|---|---|---|
| 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | 长度/bp Size | G + C/% | ||
| 1 | 黄花铁线莲 C. intricata | 550 | 60.00 | 164 | 59.15 | 227 | 65.60 |
| 2 | 芹叶铁线莲 C. aethusifolia | 549 | 60.88 | 171 | 60.36 | 219 | 67.10 |
| 3 | 褐毛铁线莲 C. fusca | 561 | 61.50 | 180 | 61.67 | 223 | 66.80 |
| 4 | 钝萼铁线莲 C. peterae | 550 | 61.27 | 172 | 60.47 | 219 | 67.60 |
| 5 | 大叶铁线莲 C. heracleifolia | 550 | 61.64 | 172 | 61.05 | 219 | 68.00 |
| 6 | 毛果扬子铁线莲 C. puberula | 543 | 60.89 | 164 | 58.90 | 220 | 67.70 |
| 7 | 狭裂太行铁线莲 C. kirilowii var. chaneti | 544 | 61.76 | 165 | 59.39 | 220 | 70.00 |
| 8 | 太行铁线莲 C. kirilowii | 545 | 61.76 | 165 | 59.76 | 221 | 69.70 |
| 9 | 短尾铁线莲 C. brevicaudata | 550 | 61.38 | 172 | 60.47 | 219 | 67.90 |
| 10 | 卷萼铁线莲 C. tubulosa | 550 | 61.57 | 172 | 61.05 | 219 | 67.90 |
| 11 | 粗齿铁线莲 C. argentilucida | 550 | 61.68 | 172 | 60.59 | 219 | 68.50 |
| 12 | 灌木铁线莲 C. fruticosa | 543 | 60.04 | 165 | 56.97 | 219 | 67.10 |
| 13 | 棉团铁线莲 C. hexapetala | 549 | 61.38 | 169 | 59.17 | 221 | 69.20 |
| 14 | 羽叶铁线莲 C. pinnata | 550 | 61.79 | 172 | 61.05 | 219 | 68.50 |
| 15 | 长瓣铁线莲 C. macropetala | 548 | 60.95 | 171 | 59.65 | 218 | 67.40 |
| 16 | 槭叶铁线莲 C. acerfolia | 559 | 62.79 | 180 | 63.89 | 220 | 68.60 |
| 17 | 半钟铁线莲 C. ochotensis | 557 | 61.40 | 179 | 60.89 | 219 | 67.60 |
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 29 | 47 | 48 | 49 | 53 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 79 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | T | A | G | C | A | G | G | C | G | — | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | T | A | G | C | G | A | G | C | G | T | C | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | T | A | G | C | A | A | G | C | G | C | C | G | C | C | C | C | G | G | C | G | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | G | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | A | R | C | G | A | G | C | G | C | T | — | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | T | T | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | T | W | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | T | A | G | Y | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | T | A | G | C | A | G | G | A | A | A | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | A | G | C | A | A | C | T | T | A | G | — | C | G | G | — | — | — | — | — | — | — | G | G | A | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | T | A | G | C | G | A | A | C | G | C | G | — | C | C | C | — | — | — | — | — | — | — | C | G | G | T | C | C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | A | G | C | A | G | G | C | G | C | C | G | C | C | T | C | C | — | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | T | A | G | C | G | C | A | C | G | C | G | — | C | C | T | C | C | G | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | C | A | C | A | A | T | T | A | C | C | G | G | T | A | G | A | A | C | T | T | T | A | T | — | C | C | C | T | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | C | T | A | W | T | T | A | C | C | K | G | T | A | G | A | A | T | T | T | T | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | C | T | — | T | G | T | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | T | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | C | T | T | T | A | C | C | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp | Loci | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | C | C | A | A | T | T | A | A | C | R | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | C | C | G | A | T | T | A | A | C | G | G | T | A | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | C | C | A | A | T | T | A | G | A | G | G | T | A | G | G | A | C | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | T | T | A | C | — | G | A | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | C | T | A | A | C | C | G | A | C | G | G | C | G | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | C | T | A | A | T | T | A | A | C | G | G | T | T | G | A | A | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 407 | 408 | 416 | 428 | 429 | 464 | 469 | 470 | 475 | 488 | 492 | 499 | 505 | 532 | 539 | 541 | 544 | 546 | 547 | 548 | 549 | 550 | 551 | 552 | 553 | 554 | 556 | 558 | 559 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | A | G | T | T | T | T | T | T | C | T | C | C | T | G | T | T | T | C | C | C | G | G | T | C | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | C | A | C | A | C | G | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | G | T | T | T | T | T | T | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | G | T | T | T | T | C | C | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | C | A | T | T | T | T | T | C | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | - | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | G | C | T | T | T | T | T | T | G | C | C | T | R | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | G | T | T | T | T | C | C | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | G | T | T | T | C | T | T | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | G | T | T | T | T | C | C | C | G | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | A | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | G | C | T | T | T | Y | Y | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | G | T | T | T | T | T | T | C | C | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | G | T | C | C | T | T | C | C | G | C | T | C | G | T | T | C | G | G | — | — | — | — | — | T | A | A | C | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | G | T | T | T | T | T | T | C | G | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
表4 17种铁线莲属植物的特异性鉴别位点
Table 4 Singletom specific discrimination site of 17 species in Clematis
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 29 | 47 | 48 | 49 | 53 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 79 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | T | A | G | C | A | G | G | C | G | — | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | T | A | G | C | G | A | G | C | G | T | C | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | T | A | G | C | A | A | G | C | G | C | C | G | C | C | C | C | G | G | C | G | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | G | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | A | R | C | G | A | G | C | G | C | T | — | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | T | T | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | T | W | G | C | A | A | C | C | C | G | G | — | T | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | T | A | G | Y | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | T | A | G | C | A | G | G | A | A | A | C | — | C | C | — | — | — | — | — | — | — | — | — | — | — | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | A | G | C | A | A | C | T | T | A | G | — | C | G | G | — | — | — | — | — | — | — | G | G | A | — | — | — | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | T | A | G | C | G | A | G | C | G | C | T | — | C | G | C | — | — | — | — | — | — | — | G | C | G | C | C | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | T | A | G | C | G | A | A | C | G | C | G | — | C | C | C | — | — | — | — | — | — | — | C | G | G | T | C | C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | A | G | C | A | G | G | C | G | C | C | G | C | C | T | C | C | — | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | T | A | G | C | G | C | A | C | G | C | G | — | C | C | T | C | C | G | G | C | G | C | G | C | G | C | T | C | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | C | A | C | A | A | T | T | A | C | C | G | G | T | A | G | A | A | C | T | T | T | A | T | — | C | C | C | T | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | C | T | A | W | T | T | A | C | C | K | G | T | A | G | A | A | T | T | T | T | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | C | T | — | T | G | T | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | T | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | T | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | C | T | T | T | A | C | C | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | C | C | A | A | T | T | A | G | A | G | T | C | A | G | A | A | G | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp | Loci | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 81 | 82 | 95 | 104 | 107 | 122 | 123 | 133 | 139 | 140 | 142 | 146 | 147 | 151 | 152 | 155 | 158 | 174 | 177 | 309 | 310 | 311 | 325 | 366 | 367 | 386 | 402 | 403 | 406 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | C | C | A | A | T | T | A | A | C | R | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | C | C | G | A | T | T | A | A | C | G | G | T | A | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | C | C | A | A | T | T | A | G | A | G | G | T | A | G | G | A | C | C | T | T | A | T | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | C | C | A | A | T | T | A | A | C | G | G | T | A | G | A | C | T | T | T | T | A | C | — | G | C | — | — | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | C | T | A | A | T | T | A | A | C | G | G | T | A | G | A | G | T | T | T | T | A | C | — | G | A | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | C | T | A | A | C | C | G | A | C | G | G | C | G | A | A | A | T | T | T | C | A | T | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | C | T | A | A | T | T | A | A | C | G | G | T | T | G | A | A | T | T | T | T | A | C | — | G | C | — | — | A | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本 编号 No. | 位点/bp Loci | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 407 | 408 | 416 | 428 | 429 | 464 | 469 | 470 | 475 | 488 | 492 | 499 | 505 | 532 | 539 | 541 | 544 | 546 | 547 | 548 | 549 | 550 | 551 | 552 | 553 | 554 | 556 | 558 | 559 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | A | G | T | T | T | T | T | T | C | T | C | C | T | G | T | T | T | C | C | C | G | G | T | C | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | C | A | C | A | C | G | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | C | G | T | T | T | T | T | T | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | C | G | T | T | T | T | C | C | T | G | C | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | C | A | T | T | T | T | T | C | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | - | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | C | G | C | T | T | T | C | C | C | C | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | C | G | C | T | T | T | T | T | T | G | C | C | T | R | T | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | C | G | T | T | T | T | C | C | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | C | G | T | T | T | C | T | T | T | G | C | C | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | C | G | T | T | T | T | C | T | C | G | C | T | T | G | T | T | C | G | — | — | — | — | — | — | G | A | A | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | C | G | T | T | T | T | C | C | C | G | C | C | T | G | C | T | C | G | G | A | — | — | — | — | G | G | A | A | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | C | G | C | T | T | T | Y | Y | T | G | Y | C | T | G | T | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | C | G | T | T | T | T | T | T | C | C | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | C | G | T | C | C | T | T | C | C | G | C | T | C | G | T | T | C | G | G | — | — | — | — | — | T | A | A | C | — | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | C | G | T | T | T | T | T | T | C | G | C | T | T | G | C | T | C | G | — | — | — | — | — | — | G | A | G | T | T | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 样本编 号No. | 遗传相似距离 Genetic distances among 17 plants in Clematis L. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| 1 | |||||||||||||||||
| 2 | 0.025 | ||||||||||||||||
| 3 | 0.029 | 0.015 | |||||||||||||||
| 4 | 0.035 | 0.021 | 0.031 | ||||||||||||||
| 5 | 0.033 | 0.015 | 0.025 | 0.006 | |||||||||||||
| 6 | 0.037 | 0.023 | 0.033 | 0.013 | 0.008 | ||||||||||||
| 7 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | |||||||||||
| 8 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | 0 | ||||||||||
| 9 | 0.035 | 0.017 | 0.027 | 0.008 | 0.002 | 0.01 | 0.037 | 0.037 | |||||||||
| 10 | 0.033 | 0.015 | 0.025 | 0.006 | 0 | 0.008 | 0.039 | 0.039 | 0.002 | ||||||||
| 11 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.008 | 0.039 | 0.039 | 0.006 | 0.004 | |||||||
| 12 | 0.031 | 0.021 | 0.029 | 0.033 | 0.029 | 0.037 | 0.045 | 0.045 | 0.031 | 0.029 | 0.033 | ||||||
| 13 | 0.041 | 0.035 | 0.045 | 0.043 | 0.039 | 0.039 | 0.023 | 0.023 | 0.041 | 0.039 | 0.039 | 0.039 | |||||
| 14 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.011 | 0.035 | 0.035 | 0.002 | 0.004 | 0.008 | 0.033 | 0.039 | ||||
| 15 | 0.041 | 0.017 | 0.025 | 0.023 | 0.017 | 0.021 | 0.037 | 0.037 | 0.019 | 0.017 | 0.017 | 0.035 | 0.041 | 0.021 | |||
| 16 | 0.049 | 0.031 | 0.039 | 0.049 | 0.043 | 0.051 | 0.057 | 0.057 | 0.045 | 0.043 | 0.047 | 0.033 | 0.055 | 0.047 | 0.045 | ||
| 17 | 0.039 | 0.015 | 0.027 | 0.023 | 0.017 | 0.021 | 0.039 | 0.039 | 0.019 | 0.017 | 0.017 | 0.031 | 0.039 | 0.021 | 0.009 | 0.039 | |
表5 ITS序列间的Kimura进化距离
Table 5 Pairwise divergence of ITS sequences using Kimura-2 parameter distance
| 样本编 号No. | 遗传相似距离 Genetic distances among 17 plants in Clematis L. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| 1 | |||||||||||||||||
| 2 | 0.025 | ||||||||||||||||
| 3 | 0.029 | 0.015 | |||||||||||||||
| 4 | 0.035 | 0.021 | 0.031 | ||||||||||||||
| 5 | 0.033 | 0.015 | 0.025 | 0.006 | |||||||||||||
| 6 | 0.037 | 0.023 | 0.033 | 0.013 | 0.008 | ||||||||||||
| 7 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | |||||||||||
| 8 | 0.045 | 0.035 | 0.045 | 0.045 | 0.039 | 0.041 | 0 | ||||||||||
| 9 | 0.035 | 0.017 | 0.027 | 0.008 | 0.002 | 0.01 | 0.037 | 0.037 | |||||||||
| 10 | 0.033 | 0.015 | 0.025 | 0.006 | 0 | 0.008 | 0.039 | 0.039 | 0.002 | ||||||||
| 11 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.008 | 0.039 | 0.039 | 0.006 | 0.004 | |||||||
| 12 | 0.031 | 0.021 | 0.029 | 0.033 | 0.029 | 0.037 | 0.045 | 0.045 | 0.031 | 0.029 | 0.033 | ||||||
| 13 | 0.041 | 0.035 | 0.045 | 0.043 | 0.039 | 0.039 | 0.023 | 0.023 | 0.041 | 0.039 | 0.039 | 0.039 | |||||
| 14 | 0.037 | 0.019 | 0.029 | 0.009 | 0.004 | 0.011 | 0.035 | 0.035 | 0.002 | 0.004 | 0.008 | 0.033 | 0.039 | ||||
| 15 | 0.041 | 0.017 | 0.025 | 0.023 | 0.017 | 0.021 | 0.037 | 0.037 | 0.019 | 0.017 | 0.017 | 0.035 | 0.041 | 0.021 | |||
| 16 | 0.049 | 0.031 | 0.039 | 0.049 | 0.043 | 0.051 | 0.057 | 0.057 | 0.045 | 0.043 | 0.047 | 0.033 | 0.055 | 0.047 | 0.045 | ||
| 17 | 0.039 | 0.015 | 0.027 | 0.023 | 0.017 | 0.021 | 0.039 | 0.039 | 0.019 | 0.017 | 0.017 | 0.031 | 0.039 | 0.021 | 0.009 | 0.039 | |
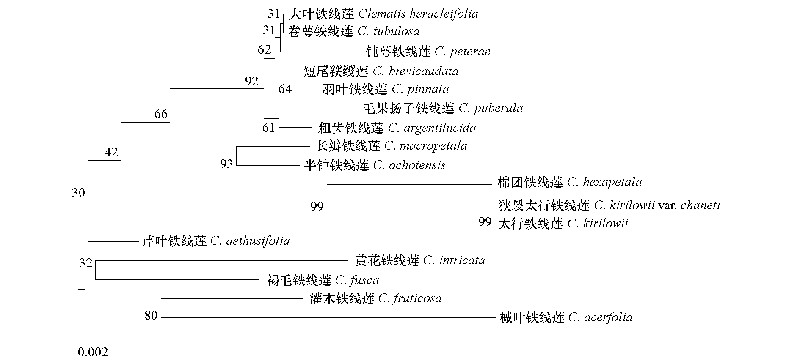
图5 基于 ITS序列构建的铁线莲系统发育树 0.002为各分支比例尺长度。
Fig. 5 Molecular phylogenetic tree inferred from ITS of 17 species in Clematis The length of the scale is 0.002,which can be used to calculate the genetic distance of each branch.
| [1] | Jiang Ming, Zhou Ying-qiao, Li Rong-rong. 2011. Sequence analysis of ITS of 8 medicinal plants of Clematis. Chinese Herbal Medicine, 42(9):1802-1806. (in Chinese) |
| 蒋明, 周英巧, 李嵘嵘. 2011. 铁线莲属8种药用植物ITS序列分析. 中草药, 42(9):1802-1806. | |
| [2] | Li Xiao, Wang Pei, Zhao Long, Ju Ai-hua. 2012. rDNA-ITS sequence analysis of Clematis mongolicum. Journal of Inner Mongolia Medical College, 34(4):325-330. (in Chinese) |
| 李骁, 王佩, 赵龙, 鞠爱华. 2012. 铁线莲属蒙药材的rDNA-ITS序列分析. 内蒙古医学院学报, 34(4):325-330. | |
| [3] | Liu Jing-jing, Gao Yi-ke. 2013. Investigation on germplasm resources of clematis in Beijing area. Heilongjiang Agricultural Science,(4):65-69. (in Chinese) |
| 刘晶晶, 高亦珂. 2013. 北京地区野生铁线莲属植物种质资源调查研究. 黑龙江农业科学,(4):65-69. | |
| [4] | Liu Qing-chao, Wang Kui-ling, Lu Wan-pei, Liu Qing-hua. 2014. Reasearch progress of Clematis in China. Journal of Plant Genetic Resource, 15(3):483-490,497. (in Chinese) |
| 刘庆超, 王奎玲, 卢婉佩, 刘庆华. 2014. 我国铁线莲属植物资源研究进展. 植物遗传资源学报, 15(3):483-490,497. | |
| [5] | Mu Lin, Xie Lei. 2011. Systematic location of Clematis acerfolia—analysis of ITS and chloroplast DNA sequences. Journal of Beijing Forestry University, 33(5):49-55. (in Chinese) |
| 穆琳, 谢磊. 2011. 槭叶铁线莲的系统位置初探——来自ITS和叶绿体DNA序列片段的分析. 北京林业大学学报, 33(5):49-55. | |
| [6] | Qian Ren-juan, Zheng Jian, Hu Qing-di, Ma Xiao-hua, Zhang Xu-Le. 2017. Research progress on ornamental germplasm resources of Clematis in China. Chinese Agricultural Science Bulletin, 33(21):75-81. (in Chinese) |
| 钱仁卷, 郑坚, 胡青荻, 马晓华, 张旭乐. 2017. 中国铁线莲属观赏种质资源研究进展. 中国农学通报, 33(21):75-81. | |
| [7] |
Salem S A, Muhammad A K, Hussein M M, Ehab H E, Muhammad A, Muhammad F. 2019. Biochemical and molecular characterization of cowpea landraces using seed storage protein and SRAP marker patterns. Saudi Journal of Biological Sciences, 26(1):74-82.
doi: 10.1016/j.sjbs.2018.09.004 URL |
| [8] | Sheng Lu. 2015. Karyotype analysis and molecular systematics of Clematis[Ph. D. Dissertation]. Nanjing:Nanjing Forestry University. |
| [9] | 盛璐. 2015. 铁线莲属植物的核型分析及分子系统学研究[博士论文]. 南京:南京林业大学. |
| [10] | Shi Ling-yan, Liu Bao-wei, Gu Xin-ying. 2019. Application of biochemical and ISSR molecular markers in identification of Auricularia auricular. Northern Horticulture, 43(9):136-141. (in Chinese) |
| 史灵燕, 刘保卫, 顾新颖. 2019. 生化和ISSR分子标记在黑木耳品种鉴定中的应用, 北方园艺, 43(9):136-141. | |
| [11] | Sun Zheng-hai, Wang Jin, Li Shi-feng, Lin Kai-wen, Hu Yi-chen, Wang Shi-jin. 2012. Establishment and optimization of ISSR-PCR reaction system for Rattan. Journal of Yunnan Agricultural University(Natural Science), 27(5):746-750. (in Chinese) |
| 孙正海, 王锦, 李世峰, 林开文, 胡祎晨, 王仕锦. 2012. 滑叶藤ISSR-PCR反应体系建立及优化. 云南农业大学学报(自然科学), 27(5):746-750. | |
| [12] | Wang Jiang-yong, Qiao Qian, Zhang Jie, Hu Feng-rong. 2020. Research progress of Clematis and application in garden. Journal of Shandong Agricultural University(Natural Science), 51(2):217-221. (in Chinese) |
| 王江勇, 乔谦, 张杰, 胡凤荣. 2020. 铁线莲属植物研究进展与园林应用. 山东农业大学学报(自然科学版), 51(2):217-221. | |
| [13] | Wang Nan, Wang Jin, Li Zong-yan, Yang Kun-mei, He Hui, Xiao Zhen-dong. 2016. ISSR-PCR reaction system optimization and primer screening of Clematis horticulture varieties. Northern Horticulture,(1):80-83. (in Chinese) |
| 王楠, 王锦, 李宗艳, 杨坤梅, 何辉, 肖振东. 2016. 铁线莲园艺品种ISSR-PCR反应体系优化与引物筛选. 北方园艺,(1):80-83. | |
| [14] | Wang W T, Xie L. 2007. A revision of Clematis sect. Tubulosae(Ranunculaceae). Acta Phytotaxonomica Suiica, 45:425-457. |
| [15] | Wang Wen-cai, Li Liang-qian. 2005. A new classification system of Clematis. Acta Botanica Taxa,(5):431-488. (in Chinese) |
| 王文采, 李良千. 2005. 铁线莲属一新分类系统. 植物分类学报,(5):431-488. | |
| [16] | Xie L, Shi J H, Li L Q. 2005. Identity of Clematis tatarinowii and C. pinnata var. ternatifolia(Ranunculaceae). Annales Botanici Fennici, 42:305-308. |
| [17] |
Xie L, Wen J, Li L Q. 2011. Phylogenetic analyses of Clematis(Ranunculaceae)based on sequences of nuclear ribosomal ITS and three plastid regions. Systematic Botany, 36:907-921.
doi: 10.1600/036364411X604921 URL |
| [18] | Yang Jiu-yan, Yang Jie, Yang Ming-bo, Ju Aihua, Qu bi. 2006. ISSR analysis of medicinal plants of the genus Caragana in Ordos Plateau. Chinese Herbal Medicine, 37(10):1562-1566. (in Chinese) |
| 杨九艳, 杨劼, 杨明博, 鞠爱华, 渠弼. 2006. 鄂尔多斯高原锦鸡儿属药用植物的ISSR分析. 中草药, 37(10):1562-1566. | |
| [19] | Zhao Huan, Wu Wei, Zheng You-liang, Pan Hong-mei, Zhai Juan-yuan. 2009. Application of ribosomal DNA ITS sequence analysis in the study of medicinal plants. Shi Zhen Chinese Medicine, 20(4):959-962. (in Chinese) |
| 赵欢, 吴卫, 郑有良, 潘红梅, 翟娟园. 2009. 核糖体DNA ITS序列分析在药用植物研究中的应用. 时珍国医国药, 20(4):959-962. | |
| [20] |
Zhu Hong, Zhu Shu-xia, Li Yong-fu, Yi Xian-gui, Duan Yi-fan, Wang Xian-rong. 2018. Leaf phenotypic variation in natural populations of Cerasus dielsiana. Chinese Journal of Plant Ecology, 42(12):1168-1178. (in Chinese)
doi: 10.17521/cjpe.2018.0196 URL |
| 朱弘, 朱淑霞, 李涌福, 伊贤贵, 段一凡, 王贤荣. 2018. 尾叶樱桃天然种群叶表型性状变异研究. 植物生态学报, 42(12):1168-1178. | |
| [21] | Zhu Yuan-di, Cao Min-ge, Xu Zheng, Wang Kun, Zhang Wen. 2014. Phylogenetic relationship between Xinjiang wild apple(Malus sieversii Roem.) and Chinese apple(Malus × domestica subsp. chinesnsis)based on ITS and matK sequences. Acta Horticulturae Sinica, 41(2):227-239. (in Chinese) |
| 朱元娣, 曹敏格, 许正, 王昆, 张文. 2014. 基于ITS和matK序列探讨新疆野苹果与中国苹果的系统演化关系. 园艺学报, 41(2):227-239. |
| [1] | 邵伟丽, 孙同冰, 刘志高, . 铁线莲新品种‘星晴’[J]. 园艺学报, 2022, 49(S1): 161-162. |
| [2] | 吴凡, 江俊浩, 卢山, 张楠, 邱帅, 魏建芬, 沈柏春. 中国绣球属种质资源及其利用研究进展[J]. 园艺学报, 2022, 49(9): 2037-2050. |
| [3] | 吕正鑫, 贺艳群, 贾东峰, 黄春辉, 钟敏, 廖光联, 朱壹, 袁开昌, 刘传浩, 徐小彪. 猕猴桃种质资源表型性状遗传多样性分析[J]. 园艺学报, 2022, 49(7): 1571-1581. |
| [4] | 李平平, 张祥, 刘雨婷, 谢志和, 张芮豪, 赵凯, 吕俊恒, 王梓然, 文锦芬, 邹学校, 邓明华. 辣椒63份种质果皮颜色与呈色物质的关系[J]. 园艺学报, 2022, 49(7): 1589-1601. |
| [5] | 张萌, 单玉莹, 杨业波, 翟飞飞, 王兆山, 巨关升, 孙振元, 李振坚. 中国石斛属植物遗传资源的AFLP分析[J]. 园艺学报, 2022, 49(6): 1339-1350. |
| [6] | 蔡志翔, 严娟, 宿子文, 徐子媛, 张明昊, 沈志军, 杨军, 马瑞娟, 俞明亮. 不同类型桃种质资源主要酚类物质含量评价[J]. 园艺学报, 2022, 49(5): 1008-1022. |
| [7] | 张世才, 李怡斐, 王春萍, 杨小苗, 黄启中, 黄任中. 辣椒种质尖孢炭疽菌抗性鉴定与评价[J]. 园艺学报, 2022, 49(4): 885-892. |
| [8] | 徐婉, 林雅君, 赵荘, 周庄. 兰属植物资源与育种研究进展[J]. 园艺学报, 2022, 49(12): 2722-2742. |
| [9] | 吴致君, 李伟, 王兴华, 张成, 蒋勋, 黎盛, 罗理勇, 孙康, 曾亮. 基于SLAF-seq的茶组子房室数不稳定居群种质亲缘关系分析[J]. 园艺学报, 2022, 49(11): 2455-2470. |
| [10] | 邓秀新. 中国柑橘育种60年回顾与展望[J]. 园艺学报, 2022, 49(10): 2063-2074. |
| [11] | 苏江硕, 贾棣文, 王思悦, 张飞, 蒋甲福, 陈素梅, 房伟民, 陈发棣. 中国菊花遗传育种60年回顾与展望[J]. 园艺学报, 2022, 49(10): 2143-2162. |
| [12] | 陈明堃, 陈璐, 孙维红, 马山虎, 兰思仁, 彭东辉, 刘仲健, 艾叶. 建兰种质资源遗传多样性分析及核心种质构建[J]. 园艺学报, 2022, 49(1): 175-186. |
| [13] | 李艳红, 聂俊, 郑锦荣, 谭德龙, 张长远, 史亮亮, 谢玉明. 华南地区樱桃番茄表型性状遗传多样性分析及综合评价[J]. 园艺学报, 2021, 48(9): 1717-1730. |
| [14] | 王力荣, 吴金龙. 中国果树种质资源研究与新品种选育70年[J]. 园艺学报, 2021, 48(4): 749-758. |
| [15] | 王振江, 罗国庆, 戴凡炜, 肖更生, 林森, 李智毅, 唐翠明. 基于8个农艺性状的569份果桑种质遗传多样性分析[J]. 园艺学报, 2021, 48(12): 2375-2384. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
版权所有 © 2012 《园艺学报》编辑部 京ICP备10030308号-2 国际联网备案号 11010802023439
编辑部地址: 北京市海淀区中关村南大街12号中国农业科学院蔬菜花卉研究所 邮编: 100081
电话: 010-82109523 E-Mail: yuanyixuebao@126.com
技术支持:北京玛格泰克科技发展有限公司